Twist transcription factor
Twist-related protein 1 (TWIST1) also known as class A basic helix–loop–helix protein 38 (bHLHa38) is a basic helix-loop-helix transcription factor that in humans is encoded by the TWIST1 gene.[5][6]
Function
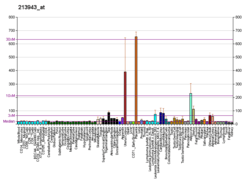
Basic helix-loop-helix (bHLH) transcription factors have been implicated in cell lineage determination and differentiation. The protein encoded by this gene is a bHLH transcription factor and shares similarity with another bHLH transcription factor, Dermo1 (a.k.a. TWIST2). The strongest expression of this mRNA is in placental tissue; in adults, mesodermally derived tissues express this mRNA preferentially.[7]
Twist1 is thought to regulate osteogenic lineage.[8]
Clinical significance
Mutations in the TWIST1 gene are associated with Saethre–Chotzen syndrome,[9][10] breast cancer,[11] and Sézary syndrome.[12]
As an oncogene
Twist plays an essential role in cancer metastasis. Over-expression of Twist or methylation of its promoter is common in metastatic carcinomas. Hence targeting Twist has a great promise as a cancer therapeutic.[13] In cooperation with N-Myc, Twist-1 acts as an oncogene in several cancers including neuroblastoma.[11][14]
Twist is activated by a variety of signal transduction pathways, including Akt, signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase, Ras, and Wnt signaling. Activated Twist upregulates N-cadherin and downregulates E-cadherin, which are the hallmarks of EMT. Moreover, Twist plays an important role in some physiological processes involved in metastasis, like angiogenesis, invadopodia, extravasation, and chromosomal instability. Twist also protects cancer cells from apoptotic cell death. In addition, Twist is responsible for the maintenance of cancer stem cells and the development of chemotherapy resistance.[13][15] Twist1 is extensively studied for its role in head- and neck cancers.[16] Here and in epithelial ovarian cancer, Twist1 has been shown to be involved in evading apoptosis, making the tumour cells resistant against platinum-based chemotherapeutic drugs like cisplatin.[15][17] Moreover, Twist1 has been shown to be expressed under conditions of hypoxia, corresponding to the observation that hypoxic cells respond less to chemotherapeutic drugs.[16]
Another process in which Twist 1 is involved is tumour metastasis. The underlying mechanism is not completely understood, but it has been implicated in the upregulation of matrix metalloproteinases[18] and inhibition of TIMP.[19]
Recently, targeting Twist has gained interest as a target for cancer therapeutics. The inactivation of Twist by small interfering RNA or chemotherapeutic approach has been demonstrated in vitro. Moreover, several inhibitors which are antagonistic to the upstream or downstream molecules of Twist signaling pathways have also been identified.[13] For example, thymoquinone, a natural product downregulates TWIST1 transcription factor to reduce epithelial to mesenchymal transition, and thus inhibits cancer metastasis in cancer cell lines and xenograft model of breast cancer in mouse [20]
Interactions
Twist transcription factor has been shown to interact with EP300,[21] TCF3[22] and PCAF.[21]
See also
References
- GRCh38: Ensembl release 89: ENSG00000122691 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000035799 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Bourgeois P, Stoetzel C, Bolcato-Bellemin AL, Mattei MG, Perrin-Schmitt F (Dec 1996). "The human H-twist gene is located at 7p21 and encodes a B-HLH protein that is 96% similar to its murine M-twist counterpart". Mammalian Genome. 7 (12): 915–7. doi:10.1007/s003359900269. PMID 8995765.
- Dollfus H, Kumaramanickavel G, Biswas P, Stoetzel C, Quillet R, Denton M, Maw M, Perrin-Schmitt F (Jul 2001). "Identification of a new TWIST mutation (7p21) with variable eyelid manifestations supports locus homogeneity of BPES at 3q22". Journal of Medical Genetics. 38 (7): 470–2. doi:10.1136/jmg.38.7.470. PMC 1757180. PMID 11474656.
- "Entrez Gene: TWIST1 twist homolog 1 (acrocephalosyndactyly 3; Saethre–Chotzen syndrome) (Drosophila)".
- Lee MS, Lowe GN, Strong DD, Wergedal JE, Glackin CA (Dec 1999). "TWIST, a basic helix–loop–helix transcription factor, can regulate the human osteogenic lineage". Journal of Cellular Biochemistry. 75 (4): 566–77. doi:10.1002/(SICI)1097-4644(19991215)75:4<566::AID-JCB3>3.0.CO;2-0. PMID 10572240.
- Kress W, Schropp C, Lieb G, Petersen B, Büsse-Ratzka M, Kunz J, Reinhart E, Schäfer WD, Sold J, Hoppe F, Pahnke J, Trusen A, Sörensen N, Krauss J, Collmann H (Jan 2006). "Saethre-Chotzen syndrome caused by TWIST 1 gene mutations: functional differentiation from Muenke coronal synostosis syndrome". European Journal of Human Genetics. 14 (1): 39–48. doi:10.1038/sj.ejhg.5201507. PMID 16251895.
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW (Jan 1997). "Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome". Nature Genetics. 15 (1): 36–41. doi:10.1038/ng0197-36. PMID 8988166.
- Martin TA, Goyal A, Watkins G, Jiang WG (Jun 2005). "Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer". Annals of Surgical Oncology. 12 (6): 488–96. doi:10.1245/ASO.2005.04.010. PMID 15864483.
- van Doorn R, Dijkman R, Vermeer MH, Out-Luiting JJ, van der Raaij-Helmer EM, Willemze R, Tensen CP (Aug 2004). "Aberrant expression of the tyrosine kinase receptor EphA4 and the transcription factor twist in Sézary syndrome identified by gene expression analysis". Cancer Research. 64 (16): 5578–86. doi:10.1158/0008-5472.CAN-04-1253. PMID 15313894.
- Khan MA, Chen HC, Zhang D, Fu J (Oct 2013). "Twist: a molecular target in cancer therapeutics". Tumour Biology. 34 (5): 2497–506. doi:10.1007/s13277-013-1002-x. PMID 23873099.
- Puisieux A, Valsesia-Wittmann S, Ansieau S (Jan 2006). "A twist for survival and cancer progression". British Journal of Cancer. 94 (1): 13–7. doi:10.1038/sj.bjc.6602876. PMC 2361066. PMID 16306876.
- Roberts CM, Tran MA, Pitruzzello MC, Wen W, Loeza J, Dellinger TH, Mor G, Glackin CA (2016). "TWIST1 drives cisplatin resistance and cell survival in an ovarian cancer model, via upregulation of GAS6, L1CAM, and Akt signalling". Scientific Reports. 6: 37652. doi:10.1038/srep37652. PMC 5120297. PMID 27876874.
- Wu KJ, Yang MH (Dec 2011). "Epithelial-mesenchymal transition and cancer stemness: the Twist1-Bmi1 connection". Bioscience Reports. 31 (6): 449–55. doi:10.1042/BSR20100114. PMID 21919891.
- Zhuo WL, Wang Y, Zhuo XL, Zhang YS, Chen ZT (May 2008). "Short interfering RNA directed against TWIST, a novel zinc finger transcription factor, increases A549 cell sensitivity to cisplatin via MAPK/mitochondrial pathway". Biochemical and Biophysical Research Communications. 369 (4): 1098–102. doi:10.1016/j.bbrc.2008.02.143. PMID 18331824.
- Zhao XL, Sun T, Che N, Sun D, Zhao N, Dong XY, Gu Q, Yao Z, Sun BC (Mar 2011). "Promotion of hepatocellular carcinoma metastasis through matrix metalloproteinase activation by epithelial-mesenchymal transition regulator Twist1". Journal of Cellular and Molecular Medicine. 15 (3): 691–700. doi:10.1111/j.1582-4934.2010.01052.x. PMC 3922390. PMID 20219012.
- Okamura H, Yoshida K, Haneji T (Jul 2009). "Negative regulation of TIMP1 is mediated by transcription factor TWIST1". International Journal of Oncology. 35 (1): 181–6. doi:10.3892/ijo_00000327. PMID 19513566.
- Khan MA, Tania M, Wei C, Mei Z, Fu S, Cheng J, Xu J, Fu J (May 2015). "Thymoquinone inhibits cancer metastasis by downregulating TWIST1 expression to reduce epithelial to mesenchymal transition". Oncotarget. 6 (23): 19580–91. doi:10.18632/oncotarget.3973. PMC 4637306. PMID 26023736.
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L (Feb 1999). "Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A". Cell. 96 (3): 405–13. doi:10.1016/S0092-8674(00)80553-X. PMID 10025406.
- El Ghouzzi V, Legeai-Mallet L, Aresta S, Benoist C, Munnich A, de Gunzburg J, Bonaventure J (Mar 2000). "Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location". Human Molecular Genetics. 9 (5): 813–9. doi:10.1093/hmg/9.5.813. PMID 10749989.
Further reading
- Seto ML, Lee SJ, Sze RW, Cunningham ML (Dec 2001). "Another TWIST on Baller-Gerold syndrome". American Journal of Medical Genetics. 104 (4): 323–30. doi:10.1002/ajmg.10065. PMID 11754069.
- Brueton LA, van Herwerden L, Chotai KA, Winter RM (Oct 1992). "The mapping of a gene for craniosynostosis: evidence for linkage of the Saethre-Chotzen syndrome to distal chromosome 7p". Journal of Medical Genetics. 29 (10): 681–5. doi:10.1136/jmg.29.10.681. PMC 1016122. PMID 1433226.
- Bianchi DW, Cirillo-Silengo M, Luzzatti L, Greenstein RM (Jun 1981). "Interstitial deletion of the short arm of chromosome 7 without craniosynostosis". Clinical Genetics. 19 (6): 456–61. doi:10.1111/j.1399-0004.1981.tb02064.x. PMID 7296937.
- Rose CS, King AA, Summers D, Palmer R, Yang S, Wilkie AO, Reardon W, Malcolm S, Winter RM (Aug 1994). "Localization of the genetic locus for Saethre-Chotzen syndrome to a 6 cM region of chromosome 7 using four cases with apparently balanced translocations at 7p21.2". Human Molecular Genetics. 3 (8): 1405–8. doi:10.1093/hmg/3.8.1405. PMID 7987323.
- Maw M, Kar B, Biswas J, Biswas P, Nancarrow D, Bridges R, Kumaramanickavel G, Denton M, Badrinath SS (Dec 1996). "Linkage of blepharophimosis syndrome in a large Indian pedigree to chromosome 7p". Human Molecular Genetics. 5 (12): 2049–54. doi:10.1093/hmg/5.12.2049. PMID 8968762.
- el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J (Jan 1997). "Mutations of the TWIST gene in the Saethre-Chotzen syndrome". Nature Genetics. 15 (1): 42–6. doi:10.1038/ng0197-42. PMID 8988167.
- Wang SM, Coljee VW, Pignolo RJ, Rotenberg MO, Cristofalo VJ, Sierra F (Mar 1997). "Cloning of the human twist gene: its expression is retained in adult mesodermally-derived tissues". Gene. 187 (1): 83–92. doi:10.1016/S0378-1119(96)00727-5. PMID 9073070.
- Krebs I, Weis I, Hudler M, Rommens JM, Roth H, Scherer SW, Tsui LC, Füchtbauer EM, Grzeschik KH, Tsuji K, Kunz J (Jul 1997). "Translocation breakpoint maps 5 kb 3' from TWIST in a patient affected with Saethre-Chotzen syndrome". Human Molecular Genetics. 6 (7): 1079–86. doi:10.1093/hmg/6.7.1079. PMID 9215678.
- Rose CS, Patel P, Reardon W, Malcolm S, Winter RM (Aug 1997). "The TWIST gene, although not disrupted in Saethre-Chotzen patients with apparently balanced translocations of 7p21, is mutated in familial and sporadic cases". Human Molecular Genetics. 6 (8): 1369–73. doi:10.1093/hmg/6.8.1369. PMID 9259286.
- Hamamori Y, Wu HY, Sartorelli V, Kedes L (Nov 1997). "The basic domain of myogenic basic helix-loop-helix (bHLH) proteins is the novel target for direct inhibition by another bHLH protein, Twist". Molecular and Cellular Biology. 17 (11): 6563–73. doi:10.1128/mcb.17.11.6563. PMC 232510. PMID 9343420.
- Gripp KW, Stolle CA, Celle L, McDonald-McGinn DM, Whitaker LA, Zackai EH (Jan 1999). "TWIST gene mutation in a patient with radial aplasia and craniosynostosis: further evidence for heterogeneity of Baller-Gerold syndrome". American Journal of Medical Genetics. 82 (2): 170–6. doi:10.1002/(SICI)1096-8628(19990115)82:2<170::AID-AJMG14>3.0.CO;2-X. PMID 9934984.
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L (Feb 1999). "Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A". Cell. 96 (3): 405–13. doi:10.1016/S0092-8674(00)80553-X. PMID 10025406.
- Kunz J, Hudler M, Fritz B (Aug 1999). "Identification of a frameshift mutation in the gene TWIST in a family affected with Robinow-Sorauf syndrome". Journal of Medical Genetics. 36 (8): 650–2. doi:10.1136/jmg.36.8.650 (inactive 2020-01-22). PMC 1762975. PMID 10465122.
- Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ (Sep 1999). "Twist is a potential oncogene that inhibits apoptosis". Genes & Development. 13 (17): 2207–17. doi:10.1101/gad.13.17.2207. PMC 317004. PMID 10485844.
- El Ghouzzi V, Legeai-Mallet L, Aresta S, Benoist C, Munnich A, de Gunzburg J, Bonaventure J (Mar 2000). "Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location". Human Molecular Genetics. 9 (5): 813–9. doi:10.1093/hmg/9.5.813. PMID 10749989.
- Lee MS, Lowe G, Flanagan S, Kuchler K, Glackin CA (Nov 2000). "Human Dermo-1 has attributes similar to twist in early bone development". Bone. 27 (5): 591–602. doi:10.1016/S8756-3282(00)00380-X. PMID 11062344.
- Dollfus H, Kumaramanickavel G, Biswas P, Stoetzel C, Quillet R, Denton M, Maw M, Perrin-Schmitt F (Jul 2001). "Identification of a new TWIST mutation (7p21) with variable eyelid manifestations supports locus homogeneity of BPES at 3q22". Journal of Medical Genetics. 38 (7): 470–2. doi:10.1136/jmg.38.7.470. PMC 1757180. PMID 11474656.
- Elanko N, Sibbring JS, Metcalfe KA, Clayton-Smith J, Donnai D, Temple IK, Wall SA, Wilkie AO (Dec 2001). "A survey of TWIST for mutations in craniosynostosis reveals a variable length polyglycine tract in asymptomatic individuals". Human Mutation. 18 (6): 535–41. doi:10.1002/humu.1230. PMID 11748846.
External links
- GeneReviews/NCBI.NIH.UW entry on Saethre–Chotzen syndrome
- Twist+transcription+factor at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.