GJD4
Gap junction delta-4 protein (GJD4), also known as connexin-40.1 (Cx40.1), is a protein that in humans is encoded by the GJD4 gene.[5]
| GJD4 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | GJD4, CX40.1, gap junction protein delta 4 | ||||||||||||||||||||||||
| External IDs | OMIM: 611922 MGI: 2444990 HomoloGene: 17692 GeneCards: GJD4 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) | |||||||||||||||||||||||||
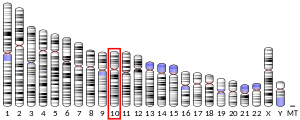
| Location (UCSC) | Chr 10: 35.61 – 35.61 Mb | Chr 18: 9.28 – 9.28 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Function
Connexins, such as GJD4, are involved in the formation of gap junctions, intercellular conduits that directly connect the cytoplasms of contacting cells. Each gap junction channel is formed by docking of 2 hemichannels, each of which contains 6 connexin subunits.[5][6]
gollark: <#348702212110680064> is implicitly self-derailed.
gollark: But probably not to geopolitics generally.
gollark: I mean, maaaaaybe strategy stuff generalizes to, say, military things somewhat.
gollark: Consider the implications for Macron.
gollark: ↑ Macron
References
- GRCh38: Ensembl release 89: ENSG00000177291 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000036855 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: gap junction protein".
- Söhl G, Nielsen PA, Eiberger J, Willecke K (2003). "Expression profiles of the novel human connexin genes hCx30.2, hCx40.1, and hCx62 differ from their putative mouse orthologues". Cell Commun. Adhes. 10 (1): 27–36. doi:10.1080/15419060302063. PMID 12881038.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.