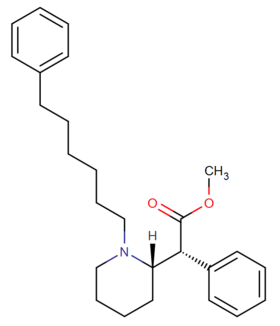
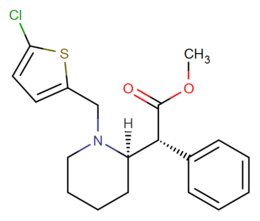
List of methylphenidate analogues
This is a list of methylphenidate (MPH or MPD) analogues, or Phenidates. The most well known compound from this family, methylphenidate, is widely prescribed around the world for the treatment of attention deficit hyperactivity disorder (ADHD) and certain other indications. Several other derivatives including rimiterol, phacetoperane and pipradrol also have more limited medical application. A rather larger number of these compounds have been sold in recent years as designer drugs, either as quasi-legal substitutes for illicit stimulants such as methamphetamine or cocaine, or as purported "study drugs" or nootropics.[1][2][3]
More structurally diverse compounds such as Desoxypipradrol (and thus Pipradrol, including such derivatives as AL-1095, Diphemethoxidine, SCH-5472 and D2PM), and even mefloquine, 2-benzylpiperidine, rimiterol, enpiroline and DMBMPP, can also be considered structurally related, with the former ones also functionally so, as loosely analogous compounds. The acyl group has sometimes been replaced with similar length ketones to increase duration. Alternatively, the methoxycarbonyl has in some cases been replaced with an alkyl group.[4][5]
Dozens more phenidates and related compounds are known from the academic and patent literature, and molecular modelling and receptor binding studies have established that the aryl and acyl substituents in the phenidate series are functionally identical to the aryl and acyl groups in the phenyltropane series of drugs, suggesting that the central core of these molecules is primarily acting merely as a scaffold to correctly orientate the binding groups, and for each of the hundreds of phenyltropanes that are known, there may be a phenidate equivalent with a comparable activity profile.
Notable phenidate derivatives
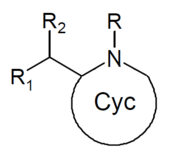
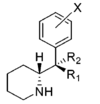
| Structure | Common name | Chemical name | CAS number | R1 | R2 |
|---|---|---|---|---|---|
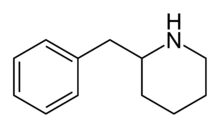 |
2-BZPD | 2-Benzylpiperidine | 32838-55-4 | phenyl | H |
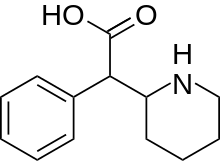 |
Ritalinic acid | Phenyl(piperidin-2-yl)acetic acid | 19395-41-6 | phenyl | COOH |
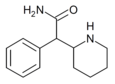 |
Ritalinamide | 2-Phenyl-2-(piperidin-2-yl)acetamide | 19395-39-2 | phenyl | CONH2 |
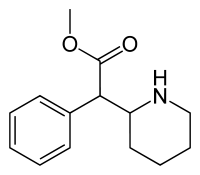 |
Methylphenidate (MPH) | Methyl phenyl(piperidin-2-yl)acetate | 113-45-1 | phenyl | COOMe |
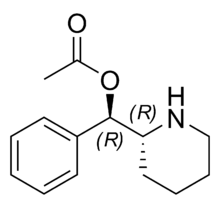 |
Phacetoperane (Lidépran) | [(R)-phenyl-[(2R)-piperidin-2-yl]methyl] acetate | 24558-01-8 | phenyl | OCOMe |
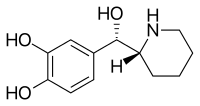 |
Rimiterol | 4-{(S)-hydroxy[(2R)-piperidin-2-yl]methyl}benzene-1,2-diol | 32953-89-2 | 3,4-dihydroxyphenyl | hydroxy |
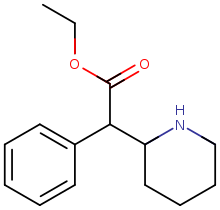 |
Ethylphenidate (EPH) | Ethyl phenyl(piperidin-2-yl)acetate | 57413-43-1 | phenyl | COOEt |
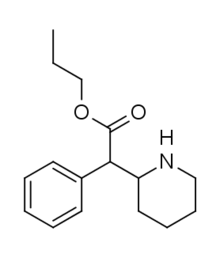 |
Propylphenidate (PPH) | Propyl phenyl(piperidin-2-yl)acetate | 1071564-47-0 | phenyl | COOnPr |
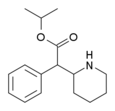 |
Isopropylphenidate (IPH) | Propan-2-yl 2-phenyl-2-(piperidin-2-yl)acetate | 93148-46-0 | phenyl | COOiPr |
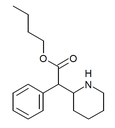 |
Butylphenidate (BPH) | Butyl phenyl(piperidin-2-yl)acetate | phenyl | COOnBu | |
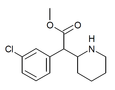 |
3-Chloromethylphenidate (3-Cl-MPH) | Methyl 2-(3-chlorophenyl)-2-(piperidin-2-yl)acetate | 191790-73-5 | 3-chlorophenyl | COOMe |
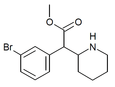 |
3-Bromomethylphenidate (3-Br-MPH) | Methyl 2-(3-bromophenyl)-2-(piperidin-2-yl)acetate | 3-bromophenyl | COOMe | |
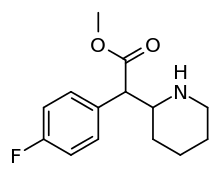 |
4-Fluoromethylphenidate (4F-MPH) | Methyl 2-(4-fluorophenyl)-2-(piperidin-2-yl)acetate | 1354631-33-6 | 4-fluorophenyl | COOMe |
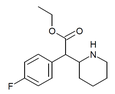 |
4-Fluoroethylphenidate (4F-EPH) | Ethyl 2-(4-fluorophenyl)-2-(piperidin-2-yl)acetate | 2160555-59-7 | 4-fluorophenyl | COOEt |
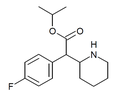 |
4-Fluoroisopropylphenidate (4F-IPH) | Propan-2-yl 2-(4-fluorophenyl)-2-(piperidin-2-yl)acetate | 4-fluorophenyl | COOiPr | |
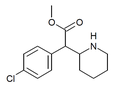 |
4-Chloromethylphenidate (4-Cl-MPH) | Methyl 2-(4-chlorophenyl)-2-(piperidin-2-yl)acetate | 680996-44-5 | 4-chlorophenyl | COOMe |
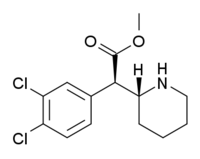 |
3,4-Dichloromethylphenidate (3,4-DCMP) | Methyl 2-(3,4-dichlorophenyl)-2-(piperidin-2-yl)acetate | 1400742-68-8 | 3,4-dichlorophenyl | COOMe |
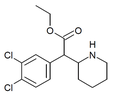 |
3,4-Dichloroethylphenidate (3,4-DCEP) | Ethyl 2-(3,4-dichlorophenyl)-2-(piperidin-2-yl)acetate | 3,4-dichlorophenyl | COOEt | |
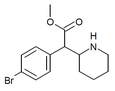 |
4-Bromomethylphenidate (4-Br-MPH) | Methyl 2-(4-bromophenyl)-2-(piperidin-2-yl)acetate | 203056-13-7 | 4-bromophenyl | COOMe |
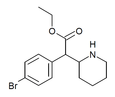 |
4-Bromoethylphenidate (4-Br-EPH) | Ethyl 2-(4-bromophenyl)-2-(piperidin-2-yl)acetate | 1391486-43-3 | 4-bromophenyl | COOEt |
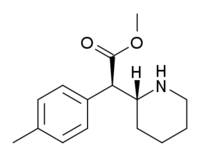 |
4-Methylmethylphenidate (4-Me-MPH) | Methyl 2-(4-methylphenyl)-2-(piperidin-2-yl)acetate | 191790-79-1 | 4-methylphenyl | COOMe |
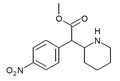 |
4-Nitromethylphenidate (4-NO2-MPH) | Methyl 2-(4-nitrophenyl)-2-(piperidin-2-yl)acetate | 4-nitrophenyl | COOMe | |
 |
Methylenedioxymethylphenidate (MDMPH) | Methyl (1,3-benzodioxol-5-yl)(piperidin-2-yl)acetate | 3,4-methylenedioxyphenyl | COOMe | |
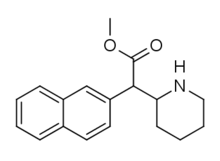 |
Methylnaphthidate (HDMP-28) | Methyl (naphthalen-2-yl)(piperidin-2-yl)acetate | 231299-82-4 | naphthalen-2-yl | COOMe |
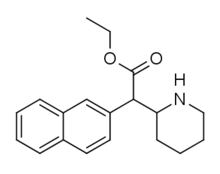 |
Ethylnaphthidate (HDEP-28) | Ethyl (naphthalen-2-yl)(piperidin-2-yl)acetate | naphthalen-2-yl | COOEt | |
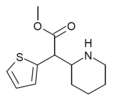 |
MTMP | Methyl (thiophen-2-yl)(piperidin-2-yl)acetate | thiophen-2-yl | COOMe | |
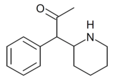 |
α-acetyl-2-benzylpiperidine | 1-Phenyl-1-(piperidin-2-yl)propan-2-one | phenyl | acetyl | |
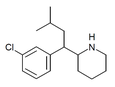 |
CPMBP | 2-[1-(3-chlorophenyl)-3-methylbutyl]piperidine | 3-chlorophenyl | isobutyl | |
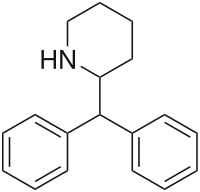 |
Desoxypipradrol (2-DPMP) | 2-benzhydrylpiperidine | 519-74-4 | phenyl | phenyl |
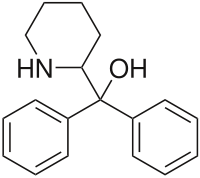 |
Pipradrol (Meratran) | Diphenyl(piperidin-2-yl)methanol | 467-60-7 | phenyl | hydroxy,phenyl |
- Related compounds
A number of related compounds are known which fit the same general structural pattern, but with substitution on the piperidine ring (e.g. SCH-5472, Difemetorex, N-benzylethylphenidate), or the piperidine ring replaced by other heterocycles such as pyrrolidine (e.g. diphenylprolinol, 2-Diphenylmethylpyrrolidine), morpholine (e.g. Methylmorphenate, 3-Benzhydrylmorpholine) or quinoline (e.g. AL-1095, Butyltolylquinuclidine).
| Structure | Common name | Chemical name | CAS number |
|---|---|---|---|
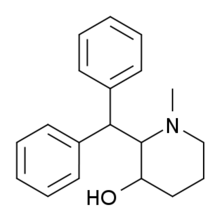 |
SCH-5472 | 2-benzhydryl-1-methyl-piperidin-3-ol | 20068-90-0 |
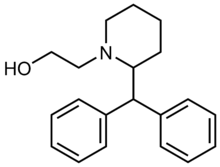 |
Difemetorex | 2-[2-(diphenylmethyl)piperidin-1-yl]ethanol | 13862-07-2 |
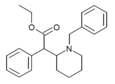 |
N-benzylethylphenidate | Ethyl (1-benzylpiperidin-2-yl)(phenyl)acetate | |
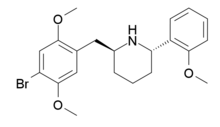 |
DMBMPP | 2-(2,5-dimethoxy-4-bromobenzyl)-6-(2-methoxyphenyl)piperidine | 1391499-52-7 |
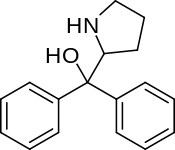 |
Diphenylprolinol (D2PM) | diphenyl(pyrrolidin-2-yl)methanol | 22348-32-9 |
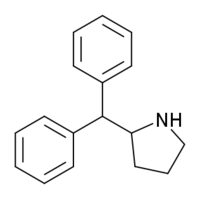 |
2-Benzhydrylpyrrolidine | 2-(Diphenylmethyl)pyrrolidine | 119237-64-8 |
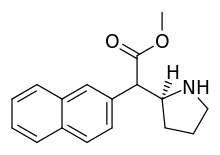 |
HDMP-29 | Methyl (naphthalen-2-yl)(pyrrolidin-2-yl)acetate | |
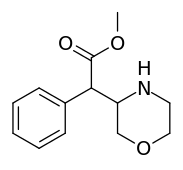 |
Methylmorphenate | Methyl morpholin-3-yl(phenyl)acetate | |
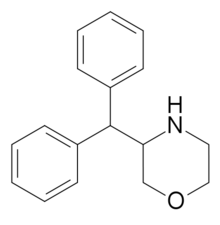 |
3-Benzhydrylmorpholine | 3-(diphenylmethyl)morpholine | 93406-27-0 |
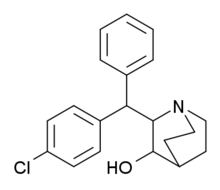 |
AL-1095 | 2-(1-phenyl-1-(p-chlorophenyl)methyl)-3-hydroxyquinuclidine | 54549-19-8 |
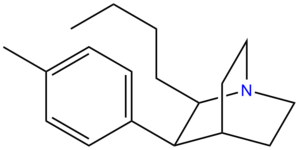 |
Butyltolylquinuclidine | (2R,3S,4S)-2-butyl-3-p-tolylquinuclidine |
Isomerism
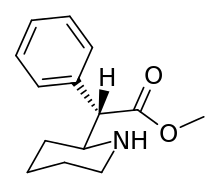
N.B. although the cyclohexane conformation, if considering both the hydrogen on the plain bond and the implicit carbon on the dotted bond are not shown as positioned as would be for the least energy state inherent to what rules apply, internally, to the molecule in and of itself: possibility of movement between putative other ligand sites in suchwise, here regarding what circumstance allows for describing it as "flexed" thus mean it has shown tendency for change in situ depending on its environment and adjacent sites of potential interaction as against its least energy state.
Methylphenidate (and its derivatives) have two chiral centers, meaning that it, and each of its analogues, have four possible enantiomers, each with differing pharmacokinetics and receptor binding profiles. In practice methylphenidate is most commonly used as pairs of diastereomers rather than isolated single enantiomers or a mixture of all four isomers. Forms include the racemate, the enantiopure (dextro or levo) of its stereoisomers; erythro or threo (either + or -) among its diastereoisomers, the chiral isomers S,S; S,R/R,S or R,R and, lastly, the isomeric conformers (which are not absolute) of either its anti- or gauche- rotamer. The variant with optimized efficacy is not the usually attested generic or common pharmaceutical brands (e.g. Ritalin, Daytrana etc.) but the (R,R)-dextro-(+)-threo-anti (sold as Focalin), which has a binding profile on par with or better than that of cocaine.[lower-alpha 1] (Note however the measure of fivefold (5×) discrepancy in the entropy of binding at their presumed shared target binding site, which may account for the higher abuse potential of cocaine over methylphenidate despite affinity for associating; i.e the latter dissociates more readily once bound despite efficacy for binding.[lower-alpha 2]) Furthermore, the energy to change between its two rotamers involves the stabilizing of the hydrogen bond between the protonated amine (of an 8.5 pKa) with the ester carbonyl resulting in reduced instances of "gauche—gauche" interactions via its favoring for activity the "anti"-conformer for putative homergic-psychostimulating pharmacokinetic properties, postulating that one inherent conformational isomer ("anti") is necessitated for the activity of the threo diastereoisomer.[lower-alpha 3]
Also of note is that methylphenidate in demethylated form is acidic; a metabolite (and precursor) known as ritalinic acid.[7] This gives the potential to yield a conjugate salt[8] form effectively protonated by a salt nearly chemically duplicate/identical to its own structure; creating a "methylphenidate ritalinate".[9]
Receptor binding profiles of selected methylphenidate analogues
Aryl substitutions
| Compound | S. Singh's alphanumeric assignation (name) |
R1 | R2 | IC50 (nM) (Inhibition of [3H]WIN 35428 binding) |
IC50 (nM) (Inhibition of [3H]DA uptake) |
Selectivity uptake/binding |
|---|---|---|---|---|---|---|
 | ||||||
| (D-threo-methylphenidate) | H, H | 33 | 244 ± 142 (171 ± 10) | 7.4 | ||
| (L-threo-methylphenidate) | 540 | 5100 (1468 ± 112) | 9.4 | |||
| (D/L-threo-methylphenidate) "eudismic ratio" | 6.4 | 20.9 (8.6) | - | |||
| (DL-threo-methylphenidate) | 83.0 ± 7.9 | 224 ± 19 | 2.7 | |||
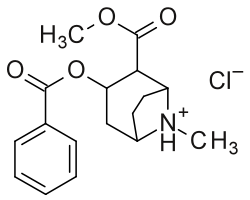 | (R-benzoyl-methylecgonine) (cocaine) | (H, H) | 173 ± 13 | 404 ± 26 | 2.3 | |
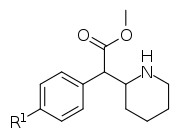 | ||||||
| 351a (4F-MPH) | F | H y d r o g e n i.e. H | 35.0 ± 3.0 | 142 ± 2.0 | 4.1 | |
| 351b | Cl | 20.6 ± 3.4 | 73.8 ± 8.1 | 3.6 | ||
| 351c | Br | 6.9 ± 0.1 | 26.3 ± 5.8 | 3.8 | ||
| 351d | (d) Br | - | 22.5 ± 2.1 | - | ||
| 351e | (l) Br | - | 408 ± 17 | - | ||
| 351d/e "eudismic ratio" | (d/l) Br | - | 18.1 | - | ||
| 351f | I | 14.0 ± 0.1 | 64.5 ± 3.5 | 4.6 | ||
| 351g | OH | 98.0 ± 10 | 340 ± 70 | 3.5 | ||
| 351h | OCH3 | 83 ± 11 | 293 ± 48 | 3.5 | ||
| 351i | (d) OCH3 | - | 205 ± 10 | - | ||
| 351j | (l) OCH3 | - | 3588 ± 310 | - | ||
| 351i/j "eudismic ratio" | (d/l) OCH3 | - | 17.5 | - | ||
| 351k (4-Me-MPH) | CH3 | 33.0 ± 1.2 | 126 ± 1 | 3.8 | ||
| 351l | t-Bu | 13500 ± 450 | 9350 ± 950 | 0.7 | ||
| 351m | NH2.HCl | 34.6 ± 4.0 | 115 ± 10 | 3.3 | ||
| 351n | NO2 | 494 ± 33 | 1610 ± 210 | 3.3 | ||
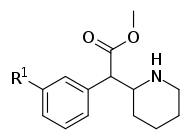 | ||||||
| 352a | F | 40.5 ± 4.5 | 160 ± 0.00 | 4.0 | ||
| 352b | Cl | 5.1 ± 1.6 | 23.0 ± 3.0 | 4.5 | ||
| 352c | Br | 4.2 ± 0.2 | 12.8 ± 0.20 | 3.1 | ||
| 352d | OH | 321 ± 1.0 | 790 ± 30 | 2.5 | ||
| 352e | OMe | 288 ± 53 | 635 ± 35 | 0.2 | ||
| 352f | Me | 21.4 ± 1.1 | 100 ± 18 | 4.7 | ||
| 352g | NH2.HCl | 265 ± 5 | 578 ± 160 | 2.2 | ||
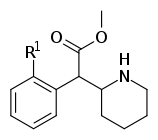 | 353a | 2′-F | 1420 ± 120 | 2900 ± 300 | 2.1 | |
| 353b | 2′-Cl | 1950 ± 230 | 2660 ± 140 | 1.4 | ||
| 353c | 2′-Br | 1870 ± 135 | 3410 ± 290 | 1.8 | ||
| 353d | 2′-OH | 23100 ± 50 | 35,800 ± 800 | 1.6 | ||
| 353e | 2′-OCH3 | 101,000 ± 10,000 | 81,000 ± 2000 | 0.8 | ||
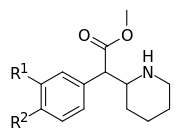 | 354a (3,4-DCMP) | Cl, Cl (3′,4′-Cl2) | 5.3 ± 0.7 | 7.0 ± 0.6 | 1.3 | |
| 354b | I | OH | 42 ± 21 | 195 ± 197 | 4.6 | |
| 354c | OMe, OMe (3′,4′-OMe2) | 810 ± 10 | 1760 ± 160 | 2.2 | ||
Both analogues 374 & 375 displayed higher potency than methylphenidate at DAT. In further comparison, 375 (the 2-naphthyl) was additionally two & a half times more potent than 374 (the 1-naphthyl isomer).[lower-alpha 5]
Aryl exchanged analogues
| Compound | S. Singh's alphanumeric assignation (name) |
Ring | Ki (nM) (Inhibition of [125I]IPT binding) |
Ki (nM) (Inhibition of [3H]DA uptake) |
Selectivity uptake/binding |
|---|---|---|---|---|---|
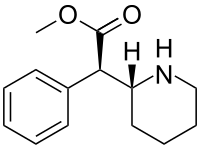 | (D-threo-methylphenidate) | benzene | 324 | - | - |
 | (DL-threo-methylphenidate) | 82 ± 77 | 429 ± 88 | 0.7 | |
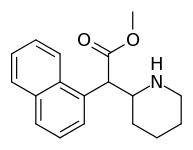 | 374 | 1-naphthalene | 194 ± 15 | 1981 ± 443 | 10.2 |
 | 375 (HDMP-28) | 2-naphthalene | 79.5 | 85.2 ± 25 | 1.0 |
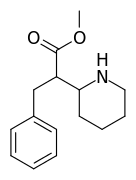 | 376 | benzyl | >5000 | - | - |

Piperidine nitrogen methylated phenyl-substituted variants
| Compound | S. Singh's alphanumeric assignation (name) |
R | IC50 (nM) (Inhibition of binding at DAT) |
|---|---|---|---|
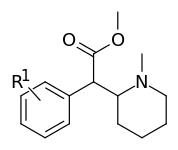 | |||
| 373a | H | 500 ± 25 | |
| 373b | 4″-OH | 1220 ± 140 | |
| 373c | 4″-CH3 | 139 ± 13 | |
| 373d | 3″-Cl | 161 ± 18 | |
| 373e | 3″-Me | 108 ± 16 |

Cycloalkane extensions, contractions & modified derivatives
| Compound | S. Singh's alphanumeric assignation (name) |
Cycloalkane ring |
Ki (nM) (Inhibition of binding) |
|---|---|---|---|
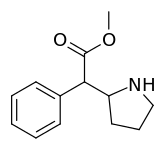 | 380 | 2-pyrrolidine (cyclopentane) | 1336 ± 108 |
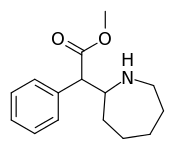 | 381 | 2-azepane (cycloheptane) | 1765 ± 113 |
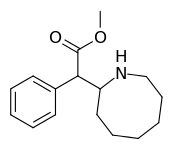 | 382 | 2-azocane (cyclooctane) | 3321 ± 551 |
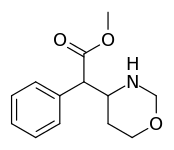 | 383 | 4-1,3-oxazinane (cyclohexane) | 6689 ± 1348 |
-2-phenylacetate.svg.png) Methyl 2-(1,2-oxazinan-3-yl)-2-phenylacetate |
-2-phenylacetate.svg.png) Methyl 2-(1,3-oxazinan-2-yl)-2-phenylacetate |
| ☝The two other (in addition to compound 383) potential oxazinane methylphenidate analogues. |
 Methyl 2-phenyl-2-(morpholin-3-yl)acetate A.K.A. Methyl 2-morpholin-3-yl-2-phenylacetate | ☜Methylmorphenate methylphenidate analogue.[11] |
Azido-iodo-N-benzyl analogues
Structures of Azido-iodo-N-benzyl analogues of methylphenidate with affinities.[12]
| Structure | Compound | R1 | R2 | Ki (nM) (Inhibition of [3H]WIN 35428 binding) |
IC50 (nM) (Inhibition of [3H]DA uptake) |
|---|---|---|---|---|---|
| (±)—threo-methylphenidate | H | H | 25 ± 1 | 156 ± 58 | |
| (±)—4-I-methylphenidate | para-iodo | H | 14 ± 3ɑ | 11 ± 2b | |
| (±)—3-I-methylphenidate | meta-iodo | H | 4.5 ± 1ɑ | 14 ± 5b | |
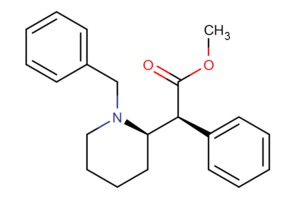 | |||||
| (±)—p-N3-N-Bn-4-I-methylphenidate | para-iodo | para-N3-N-Benzyl | 363 ± 28ɑ | 2764 ± 196bc | |
| (±)—m-N3-N-Bn-4-I-methylphenidate | para-iodo | meta-N3-N-Benzyl | 2754 ± 169ɑ | 7966 ± 348bc | |
| (±)—o-N3-N-Bn-4-I-methylphenidate | para-iodo | ortho-N3-N-Benzyl | 517 ± 65ɑ | 1232 ± 70bc | |
| (±)—p-N3-N-Bn-3-I-methylphenidate | meta-iodo | para-N3-N-Benzyl | 658 ± 70ɑ | 1828 ± 261bc | |
| (±)—m-N3-N-Bn-3-I-methylphenidate | meta-iodo | meta-N3-N-Benzyl | 2056 ± 73ɑ | 4627 ± 238bc | |
| (±)—o-N3-N-Bn-3-I-methylphenidate | meta-iodo | ortho-N3-N-Benzyl | 1112 ± 163ɑ | 2696 ± 178bc | |
| (±)—N-Bn-methylphenidate | H | N-Benzyl | — | — | |
| (±)—N-Bn-3-chloro-methylphenidate | 3-Cl | N-Benzyl | — | — | |
| (±)—N-Bn-3,4-dichloro-methylphenidate | 3,4-diCl | N-Benzyl | — | — | |
| (±)—p-chloro-N-Bn-methylphenidate | H | para-Cl-N-Benzyl | — | — | |
| (±)—p-methoxy-N-Bn-methylphenidate | H | para-OMe-N-Benzyl | — | — | |
| (±)—m-chloro-N-Bn-methylphenidate | H | meta-Cl-N-Benzyl | — | — | |
| (±)—p-nitro-N-Bn-methylphenidate | H | para-NO2-N-Benzyl | — | — |
- ɑp <0.05 versus Ki of (±)—threo-methylphenidate.
- bp <0.05 versus IC50 of (±)—threo-methylphenidate.
- cp <0.05 versus its corresponding Ki.
Alkyl substituted-carbomethoxy analogues
| Structure | R1 | R2 | R3 | Dopamine transporter Ki (nM) (Inhibition of [I125H]RTI-55 binding) |
DA uptake IC50 (nM) |
Serotonin transporter Ki (nM) (Inhibition of [I125H]RTI-55 binding) |
5HT uptake IC50 (nM) |
Norepinephrine transporter Ki (nM) (Inhibition of [I125H]RTI-55 binding) |
NE uptake IC50 (nM) |
NE/DA selectivity (binding displacement) |
NE/DA selectivity (uptake blocking) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cocaine | — ɑ | — b | — c | 500 ± 65 | 240 ± 15 | 340 ± 40 | 250 ± 40 | 500 ± 90 | 210 ± 30 | 1.0 | 0.88 |
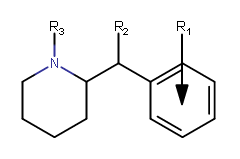 | |||||||||||
| H | COOCH3 | H | 110 ± 9 | 79 ± 16 | 65,000 ± 4,000 | 5,100 ± 7,000 | 660 ± 50 | 61 ± 14 | 6.0 | 0.77 | |
| 4-chloro | COOCH3 | H | 25 ± 8 2,000 ± 600 | 11 ± 28 2,700 ± 1,000 | 6,000 ± 100 5,900 ± 200 | >9,800 >10 mM | 110 ± 40 >6,100 | 11 ± 3 1,400 ± 400 | 4.4 | 1.0 | |
| 4-chloro | methyl | H | 180 ± 70 >3,900 | 22 ± 7 1,500 ± 700 | 4,900 ± 500 >9,100 | 1,900 ± 300 4,700 ± 800 | 360 ± 140 >6,300 | 35 ± 13 3,200 ± 800 | 2.0 | 1.6 | |
| 4-chloro | ethyl | H | 37 ± 10 1,800 ± 300 | 23 ± 5 2,800 ± 700 | 7,800 ± 800 4,200 ± 400 | 2,400 ± 400 4,100 ± 1,000 | 360 ± 60 >9,200 | 210 ± 30 1,300 ± 400 | 9.7 | 9.1 | |
| 4-chloro | propyl | H | 11 ± 3 380 ± 40 | 7.4 ± 0.4 450 ± 60 | 2,700 ± 600 3,200 ± 1,100 | 2,900 ± 1,100 1,300 ± 7 | 200 ± 80 1,400 ± 400 | 50 ± 15 200 ± 50 | 18.0 | 6.8 | |
| 4-chloro | isopropyl | H | 46 ± 16 900 ± 320 | 32 ± 6 990 ± 280 | 5,300 ± 1,300 >10 mM | 3,300 ± 400 — | 810 ± 170 >10 mM | 51 ± 20 — | 18.0 | 1.6 | |
| 4-chloro | butyl | H | 7.8 ± 1.1 290 ± 70 | 8.2 ± 2.1 170 ± 40 | 4,300 ± 400 4,800 ± 700 | 4,000 ± 400 3,300 ± 600 | 230 ± 30 1,600 ± 300 | 26 ± 7 180 ± 60 | 29.0 | 3.2 | |
| 4-chloro | isobutyl | H | 16 ± 4 170 ± 50 | 8.6 ± 2.9 380 ± 130 | 5,900 ± 900 4,300 ± 500 | 490 ± 80 540 ± 150 | 840 ± 130 4,500 ± 1,500 | 120 ± 40 750 ± 170 | 53.0 | 14.0 | |
| 4-chloro | pentyl | H | 23 ± 7 870 ± 140 | 45 ± 14 650 ± 20 | 2,200 ± 100 3,600 ± 1,000 | 1,500 ± 300 1,700 ± 700 | 160 ± 40 1,500 ± 300 | 49 ± 16 860 ± 330 | 7.0 | 1.1 | |
| 4-chloro | isopentyl | H | 3.6 ± 1.2 510 ± 170 | 14 ± 2 680 ± 120 | 5,000 ± 470 6,700 ± 500 | 7,300 ± 1,400 >8,300 | 830 ± 110 12,000 ± 1,400 | 210 ± 40 3,000 ± 540 | 230.0 | 15.0 | |
| 4-chloro | neopentyl | H | 120 ± 40 600 ± 40 | 60 ± 2 670 ± 260 | 3,900 ± 500 3,500 ± 1,000 | >8,300 1,800 ± 600 | 1,400 ± 400 >5,500 | 520 ± 110 730 ± 250 | 12.0 | 8.7 | |
| 4-chloro | cyclopentylmethyl | H | 9.4 ± 1.5 310 ± 80 | 21 ± 1 180 ± 20 | 2,900 ± 80 3,200 ± 700 | 2,100 ± 900 5,600 ± 1,400 | 1,700 ± 600 2,600 ± 800 | 310 ± 40 730 ± 230 | 180.0 | 15.0 | |
| 4-chloro | cyclohexylmethyl | H | 130 ± 40 260 ± 30 | 230 ± 70 410 ± 60 | 900 ± 400 3,700 ± 500 | 1,000 ± 200 6,400 ± 1,300 | 4,200 ± 200 4,300 ± 200 | 940 ± 140 1,700 ± 600 | 32.0 | 4.1 | |
| 4-chloro | benzyl | H | 440 ± 110 550 ± 60 | 370 ± 90 390 ± 60 | 1,100 ± 200 4,300 ± 800 | 1,100 ± 200 4,700 ± 500 | 2,900 ± 800 4,000 ± 800 | 2,900 ± 600 >8,800 | 6.6 | 7.8 | |
| 4-chloro | phenethyl | H | 24 ± 9 700 ± 90 | 160 ± 20 420 ± 140 | 640 ± 60 1,800 ± 70 | 650 ± 210 210 ± 900d | 1,800 ± 600 2,400 ± 700 | 680 ± 240 610 ± 150 | 75.0 | 4.3 | |
| 4-chloro | phenpropyl | H | 440 ± 150 2,900 ± 900 | 290 ± 90 1,400 ± 400 | 700 ± 200 1,500 ± 200 | 1,600 ± 300 1,200 ± 400 | 490 ± 100 1,500 ± 200 | 600 ± 140 1,700 ± 200 | 1.1 | 2.1 | |
| 4-chloro | 3-pentyl | H | 400 ± 80 >5,700 | 240 ± 60 1,200 ± 90 | 3,900 ± 300 4,800 ± 1,100 | >9,400 >9,600 | 970 ± 290 4,300 ± 200 | 330 ± 80 3,800 ± 30 | 2.4 | 1.4 | |
| 4-chloro | cyclopentyl | H | 36 ± 10 690 ± 140 | 27 ± 8.3 240 ± 30 | 5,700 ± 1,100 4,600 ± 700 | 4,600 ± 800 4,200 ± 900 | 380 ± 120 3,300 ± 800 | 44 ± 18 1,000 ± 300 | 11.0 | 1.6 | |
| 3-chloro | isobutyl | H | 3.7 ± 1.1 140 ± 30 | 2.8 ± 0.4 88 ± 12 | 3,200 ± 400 3,200 ± 400 | 2,100 ± 100 870 ± 230 | 23 ± 6 340 ± 50 | 14 ± 1 73 ± 5 | 6.2 | 5.0 | |
| 3,4-dichloro | COOCH3 | H | 1.4 ± 0.1 90 ± 14 | 23 ± 3 800 ± 110 | 1,600 ± 150 2,500 ± 420 | 540 ± 110 1,100 ± 90 | 14 ± 6 4,200 ± 1,900 | 10 ± 1 190 ± 50 | 10.0 | 0.43 | |
| 3,4-dichloro | propyl | H | 0.97 ± 0.31 43 ± 9 | 4.5 ± 0.4 88 ± 32 | 1,800 ± 500 450 ± 80 | 560 ± 120 180 ± 60 | 3.9 ± 1.4 30 ± 8 | 8.1 ± 3.8 47 ± 22 | 4.0 | 1.8 | |
| 3,4-dichloro | butyl | H | 2.3 ± 0.2 29 ± 5 | 5.7 ± 0.5 67 ± 13 | 1,300 ± 300 1,100 ± 200 | 1,400 ± 300 550 ± 80 | 12 ± 3 31 ± 11 | 27 ± 10 63 ± 27 | 5.2 | 4.7 | |
| 3,4-dichloro | isobutyl | H | 1.0 ± 0.5 31 ± 11 | 5.5 ± 1.3 13 ± 3 | 1,600 ± 100 450 ± 40 | 1,100 ± 300 290 ± 60 | 25 ± 9 120 ± 30 | 9.0 ± 1.2 19 ± 3 | 25.0 | 1.6 | |
| 3,4-dichloro | isobutyl | CH3 | 6.6 ± 0.9 44 ± 12 | 13 ± 4 45 ± 4 | 1,300 ± 200 1,500 ± 300 | 1,400 ± 500 2,400 ± 700 | 190 ± 60 660 ± 130 | 28 ± 3 100 ± 19 | 29.0 | 2.2 | |
| 4-methoxy | isobutyl | H | 52 ± 16 770 ± 220 | 25 ± 9 400 ± 120 | 2,800 ± 600 950 ± 190 | 3,500 ± 500 1,200 ± 300 | 3,100 ± 200 16,000 ± 2,000 | 410 ± 90 1,600 ± 400 | 60.0 | 16.0 | |
| 3-methoxy | isobutyl | H | 22 ± 5 950 ± 190 | 35 ± 12 140 ± 20 | 4,200 ± 400 3,800 ± 600 | 2,700 ± 800 2,600 ± 300 | 3,800 ± 500 12,000 ± 2,300 | 330 ± 40 1,400 ± 90 | 170.0 | 9.4 | |
| 4-isopropyl | isobutyl | H | 3,300 ± 600 >6,500 | 4,000 ± 400 >9,100 | 3,300 ± 600 1,700 ± 500 | 4,700 ± 700 1,700 ± 100 | 2,500 ± 600 3,200 ± 600 | 7,100 ± 1,800 >8,700 | 0.76 | 1.8 | |
| H | COCH3 | H | 370 ± 70 | 190 ± 50 | 7,800 ± 1,200 | >9,700 | 2,700 ± 400 | 220 ± 30 | 7.3 | 1.2 |
- ɑH = Equivalent overlay of structure sharing functional group
- bCO2CH3 (i.e. COOCH3) = Equivalent overlay of structure sharing functional group
- cCH3 = Equivalent overlay of structure sharing functional group
- dpossible typographical error in original source; e.g. 2,100 ± 900 or 900 ± 210
Restricted rotational analogs of methylphenidate (quinolizidines)
Two of the compounds tested, the weakest two @ DAT & second to the final two on the table below, were designed to elucidate the necessity of both constrained rings in the efficacy of the below series of compounds at binding by removing one or the other of the two rings in their entirety. The first of the two retain the original piperidine ring had with methylphenidate but has the constrained B ring that is common to the restricted rotational analogues thereof removed. The one below lacks the piperdine ring native to methylphenidate but keeps the ring that hindered the flexibility of the original MPH conformation. Though their potency at binding is weak in comparison to the series, with the potency shared being approximately equal between the two; the latter compound (the one more nearly resembling the substrate class of dopaminergic releasing agents similar to phenmetrazine) is 8.3-fold more potent @ DA uptake.
| Compoundɑ | R & X substitution(s) | Ki (nM) @ DAT with [33]WIN 35,065-2 |
nH @ DAT with [33]WIN 35,065-2 |
Ki (nM) or % inhibition @ NET with [33]Nisoxetine |
nH @ NET with [33]Nisoxetine |
Ki (nM) or % inhibition @ 5-HTT with [33]Citalopram |
nH @ 5-HTT with [33]Citalopram |
[33]DA uptake IC50 (nM) |
Selectivity [33]Citalopram / [33]WIN 35,065-2 |
Selectivity [33]Nisoxetine / [33]WIN 35,065-2 |
Selectivity [33]Citalopram / [33]Nisoxetine |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cocaine | — | 156 ± 11 | 1.03 ± 0.01 | 1,930 ± 360 | 0.82 ± 0.05 | 306 ± 13 | 1.12 ± 0.15 | 404 ± 26 | 2.0 | 12 | 0.16 |
| Methylphenidate | — | 74.6 ± 7.4 | 0.96 ± 0.08 | 270 ± 23 | 0.76 ± 0.06 | 14 ± 8%f | — | 230 ± 16 | >130 | 3.6 | >47 |
| 3′,4′-dichloro-MPH | — | 4.76 ± 0.62 | 2.07 ± 0.05 | NDh | — | 667 ± 83 | 1.07 ± 0.04 | 7.00 ± 140 | 140 | — | — |
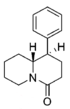 | |||||||||||
| — | 6,610 ± 440 | 0.91 ± 0.01 | 11%b | — | 3,550 ± 70 | 1.79 ± 0.55 | 8,490 ± 1,800 | 0.54 | >0.76 | <0.7 | |
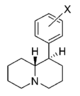 | |||||||||||
| H | 76.2 ± 3.4 | 1.05 ± 0.05 | 138 ± 9.0 | 1.12 ± 0.20 | 5,140 ± 670 | 1.29 ± 0.40 | 244 ± 2.5 | 67 | 1.8 | 37 | |
| 3′,4′-diCl | 3.39 ± 0.77 | 1.25 ± 0.29 | 28.4 ± 2.5 | 1.56 ± 0.80 | 121 ± 17 | 1.16 ± 0.31 | 11.0 ± 0.00 | 36 | 8.4 | 4.3 | |
| 2′-Cl | 480 ± 46 | 1.00 ± 0.09 | 2,750; 58%b | 0.96 | 1,840 ± 70 | 1.18 ± 0.06 | 1,260 ± 290 | 3.8 | 5.7 | 0.67 | |
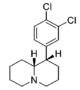 | |||||||||||
| — | 34.6 ± 7.6 | 0.95 ± 0.18 | 160 ± 18 | 1.28 ± 0.12 | 102 ± 8.2 | 1.01 ± 0.02 | 87.6 ± 0.35 | 3.0 | 4.6 | 0.64 | |
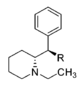 | |||||||||||
| CH2OH | 2,100 ± 697 | 0.87 ± 0.09 | NDh | — | 16.2 ± 0.05%f | — | 10,400 ± 530 | >4.8 | — | — | |
| CH3 | 7,610 ± 800 | 1.02 ± 0.03 | 8.3%b | — | 11 ± 5%f | — | 7,960 ± 290 | >1.3 | ≫0.66 | — | |
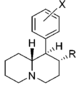 | |||||||||||
| d R=OCH3, X=H | 570 ± 49 | 0.94 ± 0.10 | 2,040; 64 ± 1.7%f | 0.73 | 14 ± 3%f | — | 1,850 ± 160 | >18 | 3.6 | >4.9 | |
| R=OH, X=H | 6,250 ± 280 | 0.86 ± 0.03 | 23.7 ± 4.1%b | — | 1 ± 1%f | — | 10,700 ± 750 | ≫1.6 | >0.80 | — | |
| R=OH, X=3′,4′-diCl | 35.7 ± 3.2 | 1.00 ± 0.09 | 367 ± 42 | 1.74 ± 0.87 | 2,050 ± 110 | 1.15 ± 0.12 | NDh | 57 | 10 | 5.6 | |
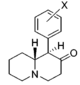 | |||||||||||
| H | 908 ± 160 | 0.88 ± 0.05 | 4030; 52%b | 1.04 | 5 ± 1%f | — | 12,400 ± 1,500 | ≫11 | 4.4 | ≫2.5 | |
| 3′,4′-diCl | 14.0 ± 1.2 | 1.27 ± 0.20 | 280 ± 76 | 0.68 ± 0.09 | 54 ± 2%f | — | NDh | ~710 | 20 | ~36 | |
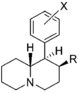 | |||||||||||
| R=OH, X=H | 108 ± 7.0 | 0.89 ± 0.10 | 351 ± 85 | 0.94 ± 0.27 | 12 ± 2%f | — | 680 ± 52 | >93 | 3.3 | >28 | |
| R=OH, X=3′,4′-diCl | 2.46 ± 0.52 | 1.39 ± 0.20 | 27.9 ± 3.5 | 0.70 ± 0.01 | 168 | 1.02 | NDh | 68 | 11 | 6.0 | |
| R=OCH3, X=H | 10.8 ± 0.8 | 0.97 ± 0.07 | 63.7 ± 2.8 | 0.84 ± 0.04 | 2,070; 73 ± 5%f | 0.90 | 61.0 ± 9.3 | 190 | 5.9 | 32 | |
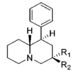 | |||||||||||
| R1=CH3, R2=H | 178 ± 28 | 1.23 ± 0.09 | 694 ± 65 | 0.88 ± 0.13 | 427 | 1.39 | 368 | 2.4 | 3.9 | 0.62 | |
| R1=H, R2=CH3 | 119 ± 20 | 1.17 ± 0.12 | 76.0 ± 12 | 0.88 ± 0.06 | 243 | 1.17 | 248 | 2.0 | 0.64 | 3.2 | |
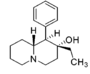 | |||||||||||
| — | 175 ± 8.0 | 1.00 ± 0.04 | 1,520 ± 120 | 0.97 ± 0.06 | 19 ± 4%f | — | NDh | >57 | 8.69 | >6.6 | |
 | |||||||||||
| R=CH2CH3, X=H | 27.6 ± 1.7 | 1.29 ± 0.05 | 441 ± 49 | 1.16 ± 0.19 | 2,390; 80%f | 1.12 | NDh | 87 | 15 | 5.8 | |
| R=CH2CH3, X=3′,4′-diCl | 3.44 ± 0.02 | 1.90 ± 0.05 | 102 ± 19 | 1.27 ± 0.10 | 286 ± 47 | 1.30 ± 0.10 | NDh | 83 | 30 | 2.8 | |
 | |||||||||||
| R=CH2CH3, X=H | 5.51 ± 0.93 | 1.15 ± 0.03 | 60.8 ± 9.6 | 0.75 ± 0.07 | 3,550; 86%f | 0.95 | NDh | 640 | 11 | 58 | |
| R=CH2CH3, X=3′,4′-diCl | 4.12 ± 0.95 | 1.57 ± 0.00 | 98.8 ± 8.7 | 1.07 ± 0.07 | 199 ± 17 | 1.24 ± 0.00 | NDh | 48 | 24 | 2.0 | |
 | |||||||||||
| — | 6,360 ± 1,300 | 1.00 ± 0.04 | 36 ± 10%c | — | 22 ± 7%f | — | 8,800 ± 870 | >1.6 | — | — | |
 | |||||||||||
| i — | 4,560 ± 1,100 | 1.10 ± 0.09 | 534 ± 210c | 0.96 ± 0.08 | 53 ± 6%f | — | 1,060 ± 115 | ~2.2 | 0.12 | ~19 | |
 | |||||||||||
| R1=CH2OH, R2=H, X=H | 406 ± 4 | 1.07 ± 0.08 | NDh | — | 31.0 ± 1.5%f | — | 1,520 ± 15 | >25 | — | — | |
| R1=CH2OCH3, R2=H, X=H | 89.9 ± 9.4 | 0.97 ± 0.04 | NDh | — | 47.8 ± 0.7%f | — | 281 ± 19 | ~110 | — | — | |
| R1=CH2OH, R2=H, X=3′,4′-diCl | 3.91 ± 0.49 | 1.21 ± 0.06 | NDh | — | 276; 94.6%f | 0.89 | 22.5 ± 1.4 | 71 | — | — | |
| R1=H, R2=CO2CH3, X=3′,4′-diCl | 363 ± 20 | 1.17 ± 0.41 | NDh | — | 2,570 ± 580 | 1.00 ± 00.1 | 317 ± 46 | 7.1 | — | — | |
| R1=CO2CH3, R2=H, X=2′-Cl | 1,740 ± 200 | 0.98 ± 0.02 | NDh | — | 22.2 ± 2.5%f | — | 2,660 ± 140 | >5.7 | — | — |
- ɑCompounds tested as hydroclhoride (HCl) salts, unless otherwise noted.
- b% inhibition caused by 5μM
- c% inhibition caused by 10μM, as assayed by SRI
- dTested as free base
- eAssayed by SRI (appropriate correction factor applied.)
- f% inhibition of 10μM compound.
- gValues expressed as x ± SEM of 2—5 replicate tests. (If no SEM shown, value is for an n of 1.)
- hNot determined
- icf. phenmetrazine & derivatives
Various MPH congener affinity values inclusive of norepinephrine & serotonin
Values for dl-threo-methylphenidate derivatives are the mean (s.d.)[16] of 3—6 determinations, or are the mean of duplicate determinations. Values of other compounds are the mean—s.d. for 3—4 determinations where indicated, or are results of single experiments which agree with the literature. All binding experiments were done in triplicate.[17]
| Compound | DA | DA Uptake | NE | 5HT |
|---|---|---|---|---|
| Methylphenidate | 84 ± 33 | 153 ± 92 | 514 ± 74 | >50,000 |
| o-Bromomethylphenidate | 880 ± 316 | — | 20,000 | — |
| m-Bromomethylphenidate | 4 ± 1 | 18 ± 11 | 20 ± 6 | 3,800 |
| p-Bromomethylphenidate | 21 ± 3 | 45 ± 19 | 31 ± 7 | 2,600 |
| p-Hydroxymethylphenidate | 125 | 263 ± 74 | 270 ± 69 | 17,000 |
| p-Methyloxymethylphenidate | 42 ± 24 | 490 ± 270 | 410 | 11,000 |
| p-Nitromethylphenidate | 180 | — | 360 | 5,900 |
| p-Iodomethylphenidate | 26 ± 14 | — | 32 | 1,800ɑ |
| m-Iodo-p-hydroxymethylphenidate | 42 ± 21 | 195 ± 197 | 370 ± 64 | 5,900 |
| N-Methylmethylphenidate | 1,400 | — | 2,800 | 40,000 |
| d-threo-Methylphenidate | 33 | — | 244 ± 142 | >50,000 |
| l-threo-Methylphenidate | 540 | — | 5,100 | >50,000 |
| dl-erythro-o-Bromomethylphenidate | 10,000 | — | 50,000 | — |
| Cocaine | 120 | 313 ± 160 | 2,100 | 190 |
| WIN 35,428 | 13 | — | 530 | 72 |
| Nomifensine | 29 ± 16 | — | 15 ± 2 | 1,300ɑ |
| Mazindol | 9 ± 5 | — | 3 ± 2 | 92 |
| Desipramine | 1,400 | — | 3.5 | 200 |
| Fluoxetine | 3,300 | — | 3,400 | 2.4 |
- ɑDenotes that preparation of membrane and results extrapolated therefrom originated from frozen tissue, which is known to change results when interpreting against fresh tissue experiments.
p-hydroxymethylphenidate displays low brain penetrability, ascribed to its phenolic hydroxyl group undergoing ionization at physiological pH.
Test environment conditioning & control studies
| Compound | 0° (zero degrees) | 0° (zero degrees) Hill slopeɑ |
22° (twenty-two degrees) | 22° (twenty-two degrees) Hill slopeɑ |
36° (thirty-six degrees) | 36° (thirty-six degrees) Hill slopeɑ |
|---|---|---|---|---|---|---|
| Methylphenidate (MPH, MPD) | 51 ± 24 | 0.99 ± 0.11 | 72 ± 29 | 0.90 ± 0.10 | 265 ± 175 | 0.70 ± 0.02 |
| o-bromo-methylphenidate | 1150 ± 83 | 0.97 ± 0.08 | 880 ± 316 | 0.79 ± 0.14 | 954 ± 190 | 0.88 ± 0.08 |
- ɑThe 'Hill' "slope" is a parameter for a biochemical equation named for Archibald Hill; 'degrees' in all cases refer to temperature, measurement of heat & cold, and not to angles. Thus "Hill slope" terminology herein has naught to do with effect of g-force or deviations of a level plane in the context of these values.
See also
piperazine.png)
References
- Klare H, Neudörfl JM, Brandt SD, Mischler E, Meier-Giebing S, Deluweit K, Westphal F, Laussmann T. Analysis of six 'neuro-enhancing' phenidate analogs. Drug Test Anal. 2017 Mar;9(3):423-435. Klare H, Neudörfl JM, Brandt SD, Mischler E, Meier-Giebing S, Deluweit K, et al. (March 2017). "Analysis of six 'neuro-enhancing' phenidate analogs" (PDF). Drug Testing and Analysis. 9 (3): 423–435. doi:10.1002/dta.2161. PMID 28067464.
- Luethi D, Kaeser PJ, Brandt SD, Krähenbühl S, Hoener MC, Liechti ME. Pharmacological profile of methylphenidate-based designer drugs. Neuropharmacology. 2018 May 15;134(Pt A):133-140. Luethi D, Kaeser PJ, Brandt SD, Krähenbühl S, Hoener MC, Liechti ME (May 2018). "Pharmacological profile of methylphenidate-based designer drugs" (PDF). Neuropharmacology. 134 (Pt A): 133–140. doi:10.1016/j.neuropharm.2017.08.020. PMID 28823611.
- Carlier J, Giorgetti R, Varì MR, Pirani F, Ricci G, Busardò FP. Use of cognitive enhancers: methylphenidate and analogs. Eur Rev Med Pharmacol Sci. 2019 Jan;23(1):3-15. Carlier J, Giorgetti R, Varì MR, Pirani F, Ricci G, Busardò FP (January 2019). "Use of cognitive enhancers: methylphenidate and analogs". European Review for Medical and Pharmacological Sciences. 23 (1): 3–15. doi:10.26355/eurrev_201901_16741. PMID 30657540.
- Froimowitz M, Gu Y, Dakin LA, Nagafuji PM, Kelley CJ, Parrish D, et al. (January 2007). "Slow-onset, long-duration, alkyl analogues of methylphenidate with enhanced selectivity for the dopamine transporter". Journal of Medicinal Chemistry. 50 (2): 219–32. doi:10.1021/jm0608614. PMID 17228864.
- Misra M, Shi Q, Ye X, Gruszecka-Kowalik E, Bu W, Liu Z, Schweri MM, Deutsch HM, Venanzi CA (2010). "Quantitative structure-activity relationship studies of threo-methylphenidate analogs". Bioorg Med Chem. 18 (20): 7221–38. doi:10.1016/j.bmc.2010.08.034. PMID 20846865.
- Singh S (March 2000). "Chemistry, design, and structure-activity relationship of cocaine antagonists" (PDF). Chemical Reviews. 100 (3): 925–1024. doi:10.1021/cr9700538. PMID 11749256.
- Marchei E, Farré M, Pardo R, Garcia-Algar O, Pellegrini M, Pacifici R, Pichini S (April 2010). "Correlation between methylphenidate and ritalinic acid concentrations in oral fluid and plasma". Clinical Chemistry. 56 (4): 585–92. doi:10.1373/clinchem.2009.138396. PMID 20167695.
- US 20040180928, Gutman A, Zaltsman I, Shalimov A, Sotrihin M, Nisnevich G, Yudovich L, Fedotev I, "Process for the preparation of dexmethylphenidate hydrochloride", published 16 September 2004, assigned to ISP Investments LLC
- US 6441178, Shahriari H, Gerard Z, Potter A, "Resolution of ritalinic acid salt", published 27 August 2002, assigned to Medeva Europe Ltd
- Lile JA, Wang Z, Woolverton WL, France JE, Gregg TC, Davies HM, Nader MA (October 2003). "The reinforcing efficacy of psychostimulants in rhesus monkeys: the role of pharmacokinetics and pharmacodynamics". The Journal of Pharmacology and Experimental Therapeutics. 307 (1): 356–66. doi:10.1124/jpet.103.049825. PMID 12954808.
- "Compound Summary for CID 85054562". PubChem. U.S. National Library of Medicine.
- Lapinsky DJ, Velagaleti R, Yarravarapu N, Liu Y, Huang Y, Surratt CK, et al. (January 2011). "Azido-iodo-N-benzyl derivatives of threo-methylphenidate (Ritalin, Concerta): Rational design, synthesis, pharmacological evaluation, and dopamine transporter photoaffinity labeling". Bioorganic & Medicinal Chemistry. 19 (1): 504–12. doi:10.1016/j.bmc.2010.11.002. PMC 3023924. PMID 21129986.
- "ChEMBL1254008". ChEMBL database. European Bioinformatics Institute (EMBL-EBI).
- "ChEMBL1255099". ChEMBL database. European Bioinformatics Institute (EMBL-EBI).
- Kim DI, Deutsch HM, Ye X, Schweri MM (May 2007). "Synthesis and pharmacology of site-specific cocaine abuse treatment agents: restricted rotation analogues of methylphenidate". Journal of Medicinal Chemistry. 50 (11): 2718–31. doi:10.1021/jm061354p. PMID 17489581.
- Jaykaran (October 2010). ""Mean ± SEM" or "Mean (SD)"?". Indian Journal of Pharmacology. 42 (5): 329. doi:10.4103/0253-7613.70402. PMC 2959222. PMID 21206631.
- Gatley SJ, Pan D, Chen R, Chaturvedi G, Ding YS (1996). "Affinities of methylphenidate derivatives for dopamine, norepinephrine and serotonin transporters". Life Sciences. 58 (12): 231–9. doi:10.1016/0024-3205(96)00052-5. PMID 8786705.
- Prinz H (March 2010). "Hill coefficients, dose-response curves and allosteric mechanisms". Journal of Chemical Biology. 3 (1): 37–44. doi:10.1007/s12154-009-0029-3. PMC 2816740. PMID 19779939.
- Endrenyi L, Fajszi C, Kwong FH (February 1975). "Evaluation of Hill slopes and Hill coefficients when the saturation binding or velocity is not known". European Journal of Biochemistry. 51 (2): 317–28. doi:10.1111/j.1432-1033.1975.tb03931.x. PMID 1149734.
- Gadagkar SR, Call GB (2015). "Computational tools for fitting the Hill equation to dose-response curves". Journal of Pharmacological and Toxicological Methods. 71: 68–76. doi:10.1016/j.vascn.2014.08.006. PMID 25157754.
Notes
- [6] ←Page #1,005 (81st page of article) §VI. Final ¶.
- [6] ←Page #1,006 (82nd page of article) 2nd column, end of first ¶.
- [6] ←Page #1,005 (81st page of article) Final § (§VI.) & page #1,006 (82nd page of article) left (1st) column, first ¶ and figure 51.
- [6] ←Page #1,010 (86th page of article) Table 47, Page #1,007 (83rd page of article) Figure 52
- [6] ←Page #1,010 (86th page of article) 2nd ¶, lines 2, 3 & 5.
- [6] ←Page #1,010 (86th page of article) Table 49, Page #1,007 (83rd page of article) Figure 54
- [6] ←Page #1,010 (86th page of article) Table 48, Page #1,007 (83rd page of article) Figure 53
- [6] ←Page #1,011 (87th page of article) Table 50, Page #1,007 (83rd page of article) Figure 55
Further reading
- Gatley SJ, Pan D, Chen R, Chaturvedi G, Ding YS (1996). "Affinities of methylphenidate derivatives for dopamine, norepinephrine and serotonin transporters". Life Sciences. 58 (12): 231–9. doi:10.1016/0024-3205(96)00052-5. PMID 8786705.
- Lapinsky DJ, Velagaleti R, Yarravarapu N, Liu Y, Huang Y, Surratt CK, et al. (January 2011). "Azido-iodo-N-benzyl derivatives of threo-methylphenidate (Ritalin, Concerta): Rational design, synthesis, pharmacological evaluation, and dopamine transporter photoaffinity labeling". Bioorganic & Medicinal Chemistry. 19 (1): 504–12. doi:10.1016/j.bmc.2010.11.002. PMC 3023924. PMID 21129986.
- Froimowitz M, Gu Y, Dakin LA, Nagafuji PM, Kelley CJ, Parrish D, et al. (January 2007). "Slow-onset, long-duration, alkyl analogues of methylphenidate with enhanced selectivity for the dopamine transporter". Journal of Medicinal Chemistry. 50 (2): 219–32. doi:10.1021/jm0608614. PMID 17228864.
- Davies HM, Hopper DW, Hansen T, Liu Q, Childers SR (April 2004). "Synthesis of methylphenidate analogues and their binding affinities at dopamine and serotonin transport sites". Bioorganic & Medicinal Chemistry Letters. 14 (7): 1799–802. doi:10.1016/j.bmcl.2003.12.097. PMID 15026075.
- Froimowitz M, Gu Y, Dakin LA, Kelley CJ, Parrish D, Deschamps JR (June 2005). "Vinylogous amide analogs of methylphenidate". Bioorganic & Medicinal Chemistry Letters. 15 (12): 3044–7. doi:10.1016/j.bmcl.2005.04.034. PMID 15908207.
- Schweri MM, Deutsch HM, Massey AT, Holtzman SG (May 2002). "Biochemical and behavioral characterization of novel methylphenidate analogs". The Journal of Pharmacology and Experimental Therapeutics. 301 (2): 527–35. doi:10.1124/jpet.301.2.527. PMID 11961053.
- Volz TJ, Bjorklund NL, Schenk JO (September 2005). "Methylphenidate analogs with behavioral differences interact differently with arginine residues on the dopamine transporter in rat striatum". Synapse. 57 (3): 175–8. doi:10.1002/syn.20161. PMID 15945061.
- Lapinsky DJ, Yarravarapu N, Nolan TL, Surratt CK, Lever JR, Tomlinson M, et al. (May 2012). "Evolution of a Compact Photoprobe for the Dopamine Transporter Based on (±)-threo-Methylphenidate". ACS Medicinal Chemistry Letters. 3 (5): 378–382. doi:10.1021/ml3000098. PMC 3469269. PMID 23066448.
- Deutsch HM, Ye X, Shi Q, Liu Z, Schweri MM (April 2001). "Synthesis and pharmacology of site specific cocaine abuse treatment agents: a new synthetic methodology for methylphenidate analogs based on the Blaise reaction". European Journal of Medicinal Chemistry. 36 (4): 303–11. doi:10.1016/s0223-5234(01)01230-2. PMID 11461755.
| Wikimedia Commons has media related to Methylphenidate. |