Deubiquitinating enzyme
Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, ubiquitin isopeptidases, are a large group of proteases[1] that cleave ubiquitin from proteins.[2] Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions.[3][4][5] DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.[2]
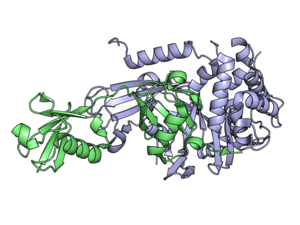
Classes
In humans there are 102 putative DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases, consisting of 58 ubiquitin-specific proteases (USPs), 4 ubiquitin C-terminal hydrolases (UCHs), 5 Machado-Josephin domain proteases (MJDs), 14 ovarian tumour proteases (OTU), and 14 Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain-containing genes. 11 of these proteins are predicted to be non-functional, leaving 79 functional enzymes.[6] In yeast, the USPs are known as ubiquitin-specific-processing proteases (UBPs).
Cysteine proteases
There are six main superfamilies of cysteine protease DUBs:[7]
- the ubiquitin-specific protease (USP/UBP) superfamily; (USP1, USP2, USP3, USP4, USP5, USP6, USP7, USP8, USP9X, USP9Y, USP10, USP11, USP12, USP13, USP14, USP15, USP16, USP17, USP17L2, USP17L3, USP17L4, USP17L5, USP17L7, USP17L8, USP18, USP19, USP20, USP21, USP22, USP23, USP24, USP25, USP26, USP27X, USP28, USP29, USP30, USP31, USP32, USP33, USP34, USP35, USP36, USP37, USP38, USP39, USP40, USP41, USP42, USP43, USP44, USP45, USP46)
- the ovarian tumour (OTU) superfamily (OTUB1, OTUB2);
- and the Machado-Josephin domain (MJD) superfamily. (ATXN3, ATXN3L)
- the ubiquitin C-terminal hydrolase (UCH) superfamily; (BAP1, UCHL1, UCHL3, UCHL5)
- the MINDY family of K48-specific deubiquitinases; (MINDY1, MINDY2, MINDY3, MINDY4)[8]
- the recently discovered ZUFSP family, at present solely represented by ZUP1[9]
| UCH | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 USP2 in complex with ubiquitin. | |||||||||
| Identifiers | |||||||||
| Symbol | UCH | ||||||||
| Pfam | PF00443 | ||||||||
| Pfam clan | CL0125 | ||||||||
| InterPro | IPR001394 | ||||||||
| PROSITE | PDOC00750 | ||||||||
| MEROPS | C19 | ||||||||
| SCOPe | 1nb8 / SUPFAM | ||||||||
| |||||||||
There is also a little known putative group of DUBs called the permutated papain fold peptidases of dsDNA viruses and eukaryote (PPPDEs) superfamily, which, if shown to be bona fide DUBs, would be the seventh in the cysteine protease class.[10]
Metalloproteases
The Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain superfamily proteins bind zinc and hence are metalloproteases.[7]
Role of deubiquitinating enzymes
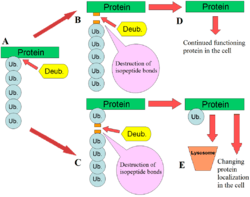
DUBs play several roles in the ubiquitin pathway. One of the best characterised functions of DUBs is the removal of monoubiqutin and polyubiquitin chains from proteins. These modifications are a post translational modification (addition to a protein after it has been made) where single ubiquitin proteins or chains of ubiquitin are added to lysines of a substrate protein. These ubiquitin modifications are added to proteins by the ubiquitination machinery; ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). The end result is ubiquitin bound to lysine residues via an isopeptide bond.[11] Proteins are affected by these modifications in a number of ways: they regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions.[3][4][5] DUBs play the antagonistic role in this axis by removing these modifications, therefore reversing the fate of the proteins.[2] In addition, a less understood role of DUBs is the cleavage of ubiquitin-like proteins such as SUMO and NEDD8. Some DUBs may have the ability to cleave isopeptide bonds between these proteins and substrate proteins.[12]
They activate ubiquitin by the proteolysis (breaking down) of the inactive expressed forms of ubiquitin. Ubiquitin is encoded in mammals by 4 different genes: UBA52, RPS27A, UBB and UBC. A similar set of genes is found in other eukaryotes such as yeast. The UBA52 and RPS27A genes produce ubiquitin that is fused to ribosomal proteins and the UBB and UBC genes produce polyubiquitin (a chain of ubiquitin joined by their C- and N-termini).[13][14] DUBs cleave the ubiquitin from these proteins, producing active single units of ubiquitin.[2]
DUBs also cleave single ubiquitin proteins that may have had their C-terminal tails accidentally bound to small cellular nucleophiles.[2] These ubiquitin-amides and ubiquitin-thioesters may be formed during standard ubiquitination reactions by the E1-E2-E3 cascade. Glutathione and polyamines are two nucleophiles that might attack the thiolester bond between ubiquitin and these enzymes. Ubiquitin C-terminal hydrolase is an example of the DUB that hydrolyses these bonds with broad specificity.[12][15]
Free polyubiquitin chains are cleaved by DUBs to produce monoubiquitin. The chains may be produced by the E1-E2-E3 machinery in the cell free from any substrate protein. Another source of free polyubiquitin is the product of ubiquitin-substrate cleavage. If DUBs cleave the base of the polyubiquitin chain that is attached to a protein, the whole chain will become free and needs to be recycled by DUBs.[2]
Domains
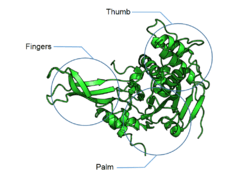
DUBs often contain a catalytic domain surrounded by one or more accessory domains, some of which contribute to target recognition. These additional domains include domain present in ubiquitin-specific proteases (DUSP) domain; ubiquitin-like (UBL) domain; meprin and TRAF homology (MATH) domain; zinc-finger ubiquitin-specific protease (ZnF-UBP) domain; zinc-finger myeloid, nervy and DEAF1 (ZnF-MYND) domain; ubiquitin-associated (UBA) domain; CHORD-SGT1 (CS) domain; microtubule-interacting and trafficking (MIT) domain; rhodenase-like domain; TBC/RABGAP domain; and B-box domain.[6][16]
Catalytic domain
The catalytic domain of DUBs is what classifies them into particular groups; USPs, OTUs, MJDs, UCHs and MPN+/JAMMs. The first 4 groups are cysteine proteases, whereas the latter is a zinc metalloprotease. The cysteine protease DUBs are papain-like and thus have a similar mechanism of action. They use either catalytic diads or triads (either two or three amino acids) to catalyse the hydrolysis of the amide bonds between ubiquitin and the substrate. The active site residues that contribute to the catalytic activity of the cysteine protease DUBs are cysteine (diad/triad), histidine (diad/triad) and aspartate or asparagine (triad only). The histidine is polarised by the aspartate or asparagine in catalytic triads or by other ways in diads. This polarised residue lowers the pKa of the cysteine, allowing it to perform a nucleophilic attack on the isopeptide bond between the ubiquitin C-terminus and the substrate lysine. Metalloproteases coordinate zinc ions with histidine, aspartate and serine residues, which activate water molecules and allows them to attack the isopeptide bond.[17][18]
UBL
Ubiquitin-like (UBL) domains have a similar structure (fold) to ubiquitin, except they lack the terminal glycine residues. 18 USPs are proposed to have UBL domains. Only 2 other DUBs have UBLs outside the USP group: OTU1 and VCPIP1. USP4, USP7, USP11, USP15, USP32, USP40 and USP47 have multiple UBL domains. Sometimes the UBL domains are in tandem, such as in USP7 where 5 tandem C-terminal UBL domains are present. USP4, USP6, USP11, USP15, USP19, USP31, USP32 and USP43 have UBL domains inserted into the catalytic domain. The functions of UBL domains are different between USPs, but commonly they regulate USP catalytic activity. They can coordinate localisation at the proteasome (USP14); negatively regulate USPs by competing for the catalytic site of the USP (USP4), and induce conformational changes to increase catalytic activity (USP7).[16][19][20] Like other UBL domains, the structure of USP UBL domains show a β-grasp fold.[21][22]
DUSP
| DUSP domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
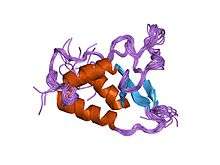 Solution structure of the DUSP domain of HUSP15. | |||||||||
| Identifiers | |||||||||
| Symbol | DUSP | ||||||||
| Pfam | PF06337 | ||||||||
| InterPro | IPR006615 | ||||||||
| MEROPS | C19 | ||||||||
| |||||||||
Single or multiple tandem DUSP domains of approximately 120 residues are found in six USPs. The function of the DUSP domain is currently unknown but it may play a role in protein-protein interaction, in particular to DUBs substrate recognition. This is predicted because of the hydrophobic cleft present in the DUSP domain of USP15 and that some protein interactions with DUSP containing USPs do not occur without these domains. The DUSP domain displays a novel tripod-like fold comprising three helices and an anti-parallel beta-sheet made of three strands. This fold resembles the legs (helices) and seat (beta-sheet) of the tripod. Within most DUSP domains in USPs there is a conserved sequence of amino acids known as the PGPI motif. This is a sequence of four amino acids; proline, glycine, proline and isoleucine, which packs against the three-helix bundle and is highly ordered.[6][23]
Role in disease
The full extent of the role of DUBs in diseases remains to be elucidated. Their involvement in disease is predicted due to known roles in physiological processes that are involved in disease states; including cancer and neurological disorders.[24]
The enzyme USP28 is over-expressed in different types of cancer such as colon or lung. In addition, USP28 deubiquitinates and stabilizes important oncogenes such as c-Myc, Notch1, c-jun or ΔNp63. [25] [26][27]
The deubiquitinating enzymes UCH-L3 and YUH1 are able to hydrolyse mutant ubiquitin UBB+1 despite of the fact that the glycine at position 76 is mutated.[28]
UCH-L1 levels are high in various types of malignancies (cancer).[29]
References
- Wilkinson KD (December 1997). "Regulation of ubiquitin-dependent processes by deubiquitinating enzymes". FASEB J. 11 (14): 1245–56. doi:10.1096/fasebj.11.14.9409543. PMID 9409543.
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009). "Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes". Annu. Rev. Biochem. 78: 363–97. doi:10.1146/annurev.biochem.78.082307.091526. PMC 2734102. PMID 19489724.
- Glickman MH, Ciechanover A (April 2002). "The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction". Physiol. Rev. 82 (2): 373–428. doi:10.1152/physrev.00027.2001. PMID 11917093.
- Mukhopadhyay D, Riezman H (January 2007). "Proteasome-independent functions of ubiquitin in endocytosis and signaling". Science. 315 (5809): 201–5. Bibcode:2007Sci...315..201M. doi:10.1126/science.1127085. PMID 17218518.
- Schnell JD, Hicke L (September 2003). "Non-traditional functions of ubiquitin and ubiquitin-binding proteins". J. Biol. Chem. 278 (38): 35857–60. doi:10.1074/jbc.R300018200. PMID 12860974.
- Nijman SM, Luna-Vargas MP, Velds A, et al. (December 2005). "A genomic and functional inventory of deubiquitinating enzymes". Cell. 123 (5): 773–86. doi:10.1016/j.cell.2005.11.007. PMID 16325574.
- Amerik AY, Hochstrasser M (November 2004). "Mechanism and function of deubiquitinating enzymes". Biochim. Biophys. Acta. 1695 (1–3): 189–207. doi:10.1016/j.bbamcr.2004.10.003. PMID 15571815.
- Abdul Rehman, Syed Arif; Kristariyanto, Yosua Adi; Choi, Soo-Youn; Nkosi, Pedro Junior; Weidlich, Simone; Labib, Karim; Hofmann, Kay; Kulathu, Yogesh (2016-07-07). "MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes". Molecular Cell. 63 (1): 146–155. doi:10.1016/j.molcel.2016.05.009. ISSN 1097-2765. PMC 4942677. PMID 27292798.
- Kwasna, Dominika; Abdul Rehman, Syed Arif; Natarajan, Jayaprakash; Matthews, Stephen; Madden, Ross; De Cesare, Virginia; Weidlich, Simone; Virdee, Satpal; Ahel, Ivan; Gibbs-Seymour, Ian; Kulathu, Yogesh (2018-04-05). "Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability". Molecular Cell. 70 (1): 150–164.e6. doi:10.1016/j.molcel.2018.02.023. ISSN 1097-2765. PMC 5896202. PMID 29576527.
- Iyer LM, Koonin EV, Aravind L (November 2004). "Novel predicted peptidases with a potential role in the ubiquitin signaling pathway". Cell Cycle. 3 (11): 1440–50. doi:10.4161/cc.3.11.1206. PMID 15483401.
- Kerscher O, Felberbaum R, Hochstrasser M (2006). "Modification of proteins by ubiquitin and ubiquitin-like proteins". Annu. Rev. Cell Dev. Biol. 22: 159–80. doi:10.1146/annurev.cellbio.22.010605.093503. PMID 16753028. S2CID 17584645.
- Wing SS (May 2003). "Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway". Int. J. Biochem. Cell Biol. 35 (5): 590–605. doi:10.1016/s1357-2725(02)00392-8. PMID 12672452.
- Kimura Y, Tanaka K (2010). "Regulatory mechanisms involved in the control of ubiquitin homeostasis". J Biochem. 147 (6): 793–8. doi:10.1093/jb/mvq044. PMID 20418328.
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A (May 1987). "The yeast ubiquitin genes: a family of natural gene fusions". EMBO J. 6 (5): 1429–39. doi:10.1002/j.1460-2075.1987.tb02384.x. PMC 553949. PMID 3038523.
- Pickart CM, Rose IA (July 1985). "Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides". J. Biol. Chem. 260 (13): 7903–10. PMID 2989266.
- Komander D, Clague MJ, Urbé S (August 2009). "Breaking the chains: structure and function of the deubiquitinases". Nat. Rev. Mol. Cell Biol. 10 (8): 550–63. doi:10.1038/nrm2731. PMID 19626045.
- Komander D (2010). Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. Subcellular Biochemistry. 54. pp. 69–87. doi:10.1007/978-1-4419-6676-6_6. ISBN 978-1-4419-6675-9. PMID 21222274.
- Chapman HA, Riese RJ, Shi GP (1997). "Emerging roles for cysteine proteases in human biology". Annu. Rev. Physiol. 59: 63–88. doi:10.1146/annurev.physiol.59.1.63. PMID 9074757.
- Faesen AC, Luna-Vargas MP, Sixma TK (June 2012). "The role of UBL domains in ubiquitin-specific proteases". Biochem. Soc. Trans. 40 (3): 539–45. doi:10.1042/BST20120004. PMID 22616864.
- Ye Y, Scheel H, Hofmann K, Komander D (December 2009). "Dissection of USP catalytic domains reveals five common insertion points". Mol Biosyst. 5 (12): 1797–808. doi:10.1039/b907669g. PMID 19734957.
- Elliott PR, Liu H, Pastok MW, et al. (November 2011). "Structural variability of the ubiquitin specific protease DUSP-UBL double domains". FEBS Lett. 585 (21): 3385–90. doi:10.1016/j.febslet.2011.09.040. PMID 22001210.
- Harper S, Besong TM, Emsley J, Scott DJ, Dreveny I (September 2011). "Structure of the USP15 N-terminal domains: a β-hairpin mediates close association between the DUSP and UBL domains". Biochemistry. 50 (37): 7995–8004. doi:10.1021/bi200726e. PMID 21848306.
- de Jong RN, Ab E, Diercks T, et al. (February 2006). "Solution structure of the human ubiquitin-specific protease 15 DUSP domain". J. Biol. Chem. 281 (8): 5026–31. doi:10.1074/jbc.M510993200. PMID 16298993.
- Singhal S, Taylor MC, Baker RT (2008). "Deubiquitylating enzymes and disease". BMC Biochem. 9 (Suppl 1): S3. doi:10.1186/1471-2091-9-S1-S3. PMC 2582804. PMID 19007433.
- E. Diefenbacher, Markus; Popov, Nikita; al., Et (June 2014). "The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer". J. Clin. Invest. 124 (8): 3407–18. doi:10.1172/JCI73733. PMC 4109555. PMID 24960159.
- Prieto-Garcia, C.; Hartmann, O; Reissland, M.; Braun, F.; Fischer, T.; Walz, S.; Fischer, A.; Calzado, M.; Orian, A.; Rosenfeldt, M.; Eilers, M.; E.Diefenbacher, M. (Jun 2019). "The USP28-∆Np63 axis is a vulnerability of squamous tumours". bioRxiv 10.1101/683508.
- Prieto-Garcia, C.; E.Diefenbacher, M.; et All (Mar 2020). "Maintaining protein stability of ∆Np63 via USP28 is required by squamous cancer cells". EMBO Mol. Med. 12 (4): e11101. doi:10.15252/emmm.201911101. PMC 7136964. PMID 32128997.
- Dennissen FJ, Kholod N, Hermes DJ, et al. (August 2011). "Mutant ubiquitin (UBB+1) associated with neurodegenerative disorders is hydrolyzed by ubiquitin C-terminal hydrolase L3 (UCH-L3)". FEBS Lett. 585 (16): 2568–74. doi:10.1016/j.febslet.2011.06.037. PMID 21762696.
- Fang Y, Fu D, Shen XZ (August 2010). "The potential role of ubiquitin c-terminal hydrolases in oncogenesis". Biochim. Biophys. Acta. 1806 (1): 1–6. doi:10.1016/j.bbcan.2010.03.001. PMID 20302916.