Isopeptide bond
An isopeptide bond is an amide bond that can form for example between the carboxyl group of one amino acid and the amino group of another. At least one of these joining groups is part of the side chain of one of these amino acids. This is unlike in a peptide bond which is sometimes called an eupeptide bond,[1] especially when discussing about both of these bond types in the same context to make a distinction between the two.
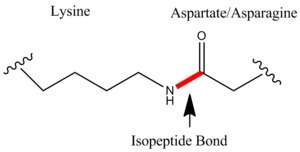
Lysine for example has an amino group on its side chain and glutamic acid has a carboxy group on its side chain. These amino acids among other similar amino acids may join together or with some other amino acids to form an isopeptide bond.
Isopeptide bond may also form between a γ-carboxamide group ( -(C=O)NH2 ) of glutamine and primary amine ( RNH2 ) of some amino acid as follows[2]
- Gln-(C=O)NH2 + RNH2 → Gln-(C=O)NH-R + NH3
Bond formation can be either enzyme catalyzed, as in the case for the isopeptide bond formed between lysine and glutamine catalyzed by transglutaminases (their reaction is similar to the reaction above),[2] or it can form spontaneously as observed in HK97 bacteriophage capsid formation[3] and Gram-positive bacterial pili.[4] Spontaneous isopeptide bond formation requires the presence of another residue, glutamic acid, which catalyzes bond formation in a proximity induced manner.[5]
An example of a small peptide containing an isopeptide bond is glutathione, which has a bond between the side chain of a glutamate residue and the amino group of a cysteine residue. An example of a protein involved in isopeptide bonding is ubiquitin, which gets attached to other proteins with a bond between the C-terminal glycine residue of ubiquitin and a lysine side chain of the substrate protein.
Biosignaling and biostructural roles
The function of enzyme generated isopeptide bonds can be roughly divided into two separate categories; signaling and structure. In the case of the former these can be a wide range of functions, influencing protein function,[6] chromatin condensation,[7] or protein half-life.[8] With regard to the latter category, isopeptides can play a role in a variety of structural aspects, from helping to form the clots in wound healing,[9] roles in extra cellular matrix upkeep & apoptosis pathway,[10] roles in the formation of pathogenic pilin,[11] restructuring of the actin skeleton of a host cell to help in the pathogenecity of V. cholerae,[12] and modifying the properties micro-tubilin to influence its role in the structure of a cell.[13]
The chemistry involved in the formation of these isopeptide bonds also tend to fall into these two categories. In the case of ubiquitin and ubiquitin-like proteins, tend to have a structured pathway of continuously passing along the peptide with a series of reactions, using multiple intermediate enzymes to reach the target protein for the conjugation reaction.[6] The structural enzymes while varying from bacterial and eukaryotic domains, tend to be single enzymes that generally in a single step, fuse the two substrates together for a larger repetitive process of linking and inter-linking the said substrates to form and influence large macromolecular structures.[14][15][12][16]
Chemistry of enzymatic bonds
Biosignaling bond chemistry
The chemistries of isopeptide bond formation are divided in the same manner as their biological roles. In the case of isopeptides used for conjugating one protein to another for the purpose of signal transduction, the literature is generally dominated by the very well-studied Ubiquitin protein and related proteins. While there are many related proteins to Ubiquitin, such as SUMO, Atg8, Atg12, and so on, they all tend to follow relatively the same protein ligation pathway.[6]
Therefore, the best example is to look at Ubiquitin, as while there can be certain differences, Ubiquitin is essentially the model followed in all these cases. The process essentially has three tiers, in the initial step, the activating protein generally denominated as E1 activates the Ubiquitin protein by adenylating it with ATP. Then the adenylated Ubiquitin is essentially activated and can be transferred to a conserved cysteine using a thioester bond which is between the carboxyl group of the c-terminal glycine of the ubiquitin and the sulfur of the E1 cysteine.[6][8] The activating E1 enzyme then binds with and transfers the Ubiquitin to the next tier, the E2 enzyme which accepts the protein and once again forms a thioester with a conserved bond. The E2 acts to certain degree as an intermediary which then binds to E3 enzyme ligase for the final tier, which leads to the eventual transfer of the ubiquitin or ubiquitin related protein to a lysine site on the targeted protein, or more commonly for ubiquitin, onto ubiquitin itself to form chains of said protein.[6]
However, in final tier, there is also a divergence, in that depending on the type of E3 ligase, it may not actually be causing the conjugation. As there are the E3 ligases containing HECT domains, in which they continue this ‘transfer chain’ by accepting once again the ubiquitin via another conserved cysteine and then targeting it and transferring it to the desired target. Yet in case of RING finger domain containing that use coordination bonds with Zinc ions to stabilize their structures, they act more to direct the reaction. By that, it's meant that once the RING finger E3 ligase binds with the E2 containing the ubiquitin, it simply acts as a targeting device which directs the E2 to directly ligate the target protein at the lysine site.[6][17]
Though in this case ubiquitin does represent other proteins related to it well, each protein obviously will have its own nuisances such as SUMO, which tends to be RING finger domain domainated ligases, where the E3 simply acts as the targeting device to direct the ligation by the E2, and not actually performing the reaction itself such as the Ubiquitin E3-HECT ligases.[8] Thus while the internal mechanisms differ such as how proteins participate in the transfer chain, the general chemical aspects such as using thioesters and specific ligases for targeting remain the same.
Biostructural bond chemistry
The enzymatic chemistry involved in the formation of isopeptides for structural purposes is different from the case of ubiquitin and ubiquitin related proteins. In that, instead of sequential steps involving multiple enzymes to activate, conjugate and target the substrate.[18] The catalysis is performed by one enzyme and the only precursor step, if there is one, is generally cleavage to activate it from a zymogen. However, the uniformity that exists in the ubiquitin’s case is not so here, as there are numerous different enzymes all performing the reaction of forming the isopeptide bond.
The first case is that of the sortases, an enzyme family that is spread throughout numerous gram positive bacteria. It has been shown to be an important pathogenicity and virulence factor. The general reaction performed by sortases involves using its own brand of the ‘catalytic triad’: i.e. using histidine, arginine, and cysteine for the reactive mechanism. His and Arg act to help create the reactive environment, and Cys once again acts as the reaction center by using a thioester help hold a carboxyl group until the amine of a Lysine can perform a nucleophilic attack to transfer the protein and form the isopeptide bond. An ion that can sometimes play an important although indirect role in the enzymatic reaction is calcium, which is bound by sortase. It plays an important role in holding the structure of the enzyme in the optimal conformation for catalysis. However, there are cases where calcium has been shown to be non-essential for catalysis to take place.[15]
Another aspect that distinguishes sortases in general is that they have a very specific targeting for their substrate, as sortases have generally two functions, the first is the fusing of proteins to the cell wall of the bacteria and the second is the polymerization of pilin. For the process of localization of proteins to the cell wall there is three-fold requirement that the protein contain a hydrophobic domain, a positively charged tail region, and final specific sequence used for recognition.[19] The best studied of these signals is the LPXTG, which acts as the point of cleavage, where the sortase attacks in between Thr and Gly, conjugating to the Thr carboxyl group.[15] Then the thioester is resolved by the transfer of the peptide to a primary amine, and this generally has a very high specificity, which is seen in the example of B. cereus where the sortase D enzyme helps to polymerize the BcpA protein via two recognition signals, the LPXTG as the cleavage and thioester forming point, and the YPKN site which acts as the recognition signal as where the isopeptide will form.[20] While the particulars may vary between bacteria, the fundamentals of sortase enzymatic chemistry remain the same.
The next case is that of Transglutaminases (TGases), which act mainly within eukaryotes for fusing together different proteins for a variety of reasons such as a wound healing or attaching proteins to lipid membranes.[21][9] The TGases themselves also contain their own ‘catalytic triad’ with Histidine, Aspartate, and Cysteine. The roles of these residues are analogous or the same as the previously described Sortases, in that His and Asp play a supporting role in interacting with the target residue, while the Cys forms a thioester with a carboxyl group for a later nucleophilic attack by a primary amine, in this case due to interest that of Lysine. Though the similarities to sortase catalytically start to end there, as the enzyme and the family is dependent on calcium, which plays a crucial structural role in holding a tight conformation of the enzyme. The TGases, also have a very different substrate specificity in that they target specifically the middle Gln, in the sequence ‘Gln-Gln-Val’. The general substrate specificity, i.e. the specific protein is due to the general structure of different TGases which targets them to the substrate.[22]
The specificity has been noted in TGases such that different TGases will react with different Gln’s on the same protein, signifying that the enzymes have a very specific initial targeting.[23] It has also been shown to have some specificity as to which target Lysine it transfers the protein to, as in the case of Factor XIII, where the adjacent residue to the Lys decides whether the reaction will occur.[9] Thus while the TGases may initially seem like a eukaryotic sortase, they stand on their own as separate set of enzymes.
Another case of an isopeptide linking enzyme for structural purposes is the actin cross-linking domain (ACD) of the MARTX toxin protein generated by V. cholerae. While it has been shown that the ACD when performing the catalysis uses magnesium and ATP for the formation of the cross-links the specifics of the mechanism are uncertain. Though an interesting aspect of the cross-link formed in this case, is that it uses a non-terminal Glu to ligate to a non-terminal Lys, which seems to be rare in the process of forming an isopeptide bond.[12] Though the chemistry of ACD is still to be resolved, it shows that isopeptide bond formation is not dependent simply on Asp/Asn for non-terminal isopeptide linkages between proteins.
The final case to be looked is the curious case of the post translational modifications of microtubilin (MT). MT contains a wide array of post translational modifications; however the two of most regarded interest are polyglutamylation and polyglycylation. Both modifications are similar in the sense they are repeating stretches of the same amino acid fused to the side chain carboxyl group of glutamate at the c-terminal region of the MT. The enzymatic mechanisms are not fully fleshed out as not much is known about the polyglycating enzyme. In the case of polyglutamylation the exact mechanism is also unknown, but it does seem to be ATP-dependent.[24] Though again there is a lack of clarity in regard to the enzymatic chemistry, there is still valuable insight in the formation of isopeptide bonds using the R-group carboxyl of Glu in conjunction with the N-terminal amino of the modifying peptides.
Spontaneous formation
Researchers have exploited spontaneous isopeptide bond formation to develop a peptide tag called SpyTag. SpyTag can spontaneously and irreversibly react with its binding partner (a protein termed SpyCatcher) through a covalent isopeptide bond.[5] This molecular tool may have applications for in vivo protein targeting, fluorescent microscopy, and irreversible attachment for a protein microarray. Following this, other Tag/Catcher systems were developed such as SnoopTag/SnoopCatcher[25] and SdyTag/SdyCatcher[26] that complement SpyTag/SpyCatcher.
See also
- Organic Chemistry
- Biochemistry
- Protein tags
- SpyCatcher
References
- "Nomenclature and Symbolism for Amino Acids and Peptides. Recommendations 1983". European Journal of Biochemistry. 138 (1): 9–37. 1984. doi:10.1111/j.1432-1033.1984.tb07877.x. ISSN 0014-2956. PMID 6692818.
- DeJong, GAH; Koppelman, SJ (2002). "Transglutaminase Catalyzed Reactions: Impact on Food Applications". Journal of Food Science. 67 (8): 2798–2806. doi:10.1111/j.1365-2621.2002.tb08819.x. ISSN 0022-1147.
- Wikoff, WR; et al. (2000). "Topologically linked protein rings in the bacteriophage HK97 capsid". Science. 289 (5487): 2129–2133. Bibcode:2000Sci...289.2129W. doi:10.1126/science.289.5487.2129.
- Kang, H. J.; Coulibaly, F.; Clow, F.; Proft, T.; Baker, E. N. (2007). "Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure". Science. 318 (5856): 1625–1628. Bibcode:2007Sci...318.1625K. doi:10.1126/science.1145806. PMID 18063798.
- Zakeri, B. (2012). "Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin". Proceedings of the National Academy of Sciences. 109 (12): E690–7. Bibcode:2012PNAS..109E.690Z. doi:10.1073/pnas.1115485109. PMC 3311370. PMID 22366317.
- Kerscher, O; Felberbaum, R; Hochstrasser, M (2006). "Modification of proteins by ubiquitin and ubiquitin-like proteins". Annual Review of Cell and Developmental Biology. 22: 159–80. doi:10.1146/annurev.cellbio.22.010605.093503. PMID 16753028.
- Turner, BM (Nov 1, 2002). "Cellular memory and the histone code". Cell. 111 (3): 285–91. doi:10.1016/S0092-8674(02)01080-2. PMID 12419240.
- Gill, G (Sep 1, 2004). "SUMO and ubiquitin in the nucleus: different functions, similar mechanisms?". Genes & Development. 18 (17): 2046–59. doi:10.1101/gad.1214604. PMID 15342487.
- Ariëns, RA; Lai, TS; Weisel, JW; Greenberg, CS; Grant, PJ (Aug 1, 2002). "Role of factor XIII in fibrin clot formation and effects of genetic polymorphisms". Blood. 100 (3): 743–54. doi:10.1182/blood.v100.3.743. PMID 12130481.
- GRIFFIN, Martin; CASADIO, Rita; BERGAMINI, Carlo M. (2002). "Transglutaminases: Nature's biological glues". Biochemical Journal. 368 (2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- Marraffini, LA; Dedent, AC; Schneewind, O (March 2006). "Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria". Microbiology and Molecular Biology Reviews. 70 (1): 192–221. doi:10.1128/MMBR.70.1.192-221.2006. PMC 1393253. PMID 16524923.
- Kudryashov, DS; Durer, ZA; Ytterberg, AJ; Sawaya, MR; Pashkov, I; Prochazkova, K; Yeates, TO; Loo, RR; Loo, JA; Satchell, KJ; Reisler, E (Nov 25, 2008). "Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin". Proceedings of the National Academy of Sciences of the United States of America. 105 (47): 18537–42. Bibcode:2008PNAS..10518537K. doi:10.1073/pnas.0808082105. PMC 2587553. PMID 19015515.
- Westermann, Stefan; Weber, Klaus (1 December 2003). "Post-translational modifications regulate microtubule function" (PDF). Nature Reviews Molecular Cell Biology. 4 (12): 938–948. doi:10.1038/nrm1260. hdl:11858/00-001M-0000-0012-EF93-5. PMID 14685172.
- Griffin, Martin; Casadio, Rita; Bergamini, Carlo M. (2002). "Transglutaminases: Nature's biological glues". Biochemical Journal. 368 (2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- Clancy, Kathleen W.; Melvin, Jeffrey A.; McCafferty, Dewey G. (30 June 2010). "Sortase transpeptidases: Insights into mechanism, substrate specificity, and inhibition". Biopolymers. 94 (4): 385–396. doi:10.1002/bip.21472. PMC 4648256. PMID 20593474.
- Westermann, Stefan; Weber, Klaus (1 December 2003). "Post-translational modifications regulate microtubule function" (PDF). Nature Reviews Molecular Cell Biology. 4 (12): 938–948. doi:10.1038/nrm1260. hdl:11858/00-001M-0000-0012-EF93-5. PMID 14685172.
- Jackson, PK; Eldridge, AG; Freed, E; Furstenthal, L; Hsu, JY; Kaiser, BK; Reimann, JD (October 2000). "The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases". Trends in Cell Biology. 10 (10): 429–39. doi:10.1016/S0962-8924(00)01834-1. PMID 10998601.
- Grabbe, C; Dikic, I (April 2009). "Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins". Chemical Reviews. 109 (4): 1481–94. doi:10.1021/cr800413p. PMID 19253967.
- Marraffini, LA; Dedent, AC; Schneewind, O (March 2006). "Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria". Microbiology and Molecular Biology Reviews. 70 (1): 192–221. doi:10.1128/MMBR.70.1.192-221.2006. PMC 1393253. PMID 16524923.
- Budzik, JM; Marraffini, LA; Souda, P; Whitelegge, JP; Faull, KF; Schneewind, O (Jul 22, 2008). "Amide bonds assemble pili on the surface of bacilli". Proceedings of the National Academy of Sciences of the United States of America. 105 (29): 10215–20. Bibcode:2008PNAS..10510215B. doi:10.1073/pnas.0803565105. PMC 2481347. PMID 18621716.
- Ahvazi, B; Steinert, PM (Aug 31, 2003). "A model for the reaction mechanism of the transglutaminase 3 enzyme". Experimental & Molecular Medicine. 35 (4): 228–42. doi:10.1038/emm.2003.31. PMID 14508061.
- Ahvazi, B; Steinert, PM (Aug 31, 2003). "A model for the reaction mechanism of the transglutaminase 3 enzyme". Experimental & Molecular Medicine. 35 (4): 228–42. doi:10.1038/emm.2003.31. PMID 14508061.
- Griffin, M; Casadio, R; Bergamini, CM (Dec 1, 2002). "Transglutaminases: nature's biological glues". The Biochemical Journal. 368 (Pt 2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- Westermann, Stefan; Weber, Klaus (1 December 2003). "Post-translational modifications regulate microtubule function" (PDF). Nature Reviews Molecular Cell Biology. 4 (12): 938–948. doi:10.1038/nrm1260. hdl:11858/00-001M-0000-0012-EF93-5. PMID 14685172.
- Veggiani, Gianluca; Nakamura, Tomohiko; Brenner, Michael D.; Gayet, Raphaël V.; Yan, Jun; Robinson, Carol V.; Howarth, Mark (2 February 2016). "Programmable polyproteams built using twin peptide superglues". Proceedings of the National Academy of Sciences. 113 (5): 1202–1207. Bibcode:2016PNAS..113.1202V. doi:10.1073/pnas.1519214113. PMC 4747704. PMID 26787909.
- Tan, Lee Ling; Hoon, Shawn S.; Wong, Fong T.; Ahmed, S. Ashraf (26 October 2016). "Kinetic Controlled Tag-Catcher Interactions for Directed Covalent Protein Assembly". PLOS ONE. 11 (10): e0165074. Bibcode:2016PLoSO..1165074T. doi:10.1371/journal.pone.0165074. PMC 5082641. PMID 27783674.