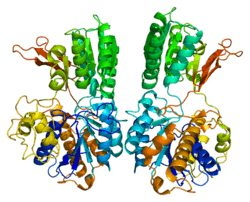
Metabotropic glutamate receptor 1
The glutamate receptor, metabotropic 1, also known as GRM1, is a human gene which encodes the metabotropic glutamate receptor 1 (mGluR1) protein.[5][6][7]
Function
L-glutamate is the major excitatory neurotransmitter in the central nervous system and activates both ionotropic and metabotropic glutamate receptors. Glutamatergic neurotransmission is involved in most aspects of normal brain function and can be perturbed in many neuropathologic conditions. The metabotropic glutamate receptors are a family of G protein-coupled receptors, that have been divided into 3 groups on the basis of sequence homology, putative signal transduction mechanisms, and pharmacologic properties. Group I, which includes GRM1 alongside GRM5, have been shown to activate phospholipase C. Group II includes GRM2 and GRM3 while Group III includes GRM4, GRM6, GRM7 and GRM8. Group II and III receptors are linked to the inhibition of the cyclic AMP cascade but differ in their agonist selectivities. Alternative splice variants of the GRM1 gene have been described but their full-length nature has not been determined.[5]
A possible connection has been suggested between mGluRs and neuromodulators, as mGluR1 antagonists block adrenergic receptor activation in neurons.[8]
Studies with knockout mice
Mice lacking functional glutamate receptor 1 were reported in 1994. By homologous recombination mediated gene targeting those mice became deficient in mGlu receptor 1 protein. The mice did not show any basic anatomical changes in the brain but had impaired cerebellar long-term depression and hippocampal long-term potentiation. In addition they had impaired motor functions, characterized by impaired balance. In the Morris watermaze test, an assay for learning abilities, those mice needed significantly more time to successfully complete the task.[9]
Ligands
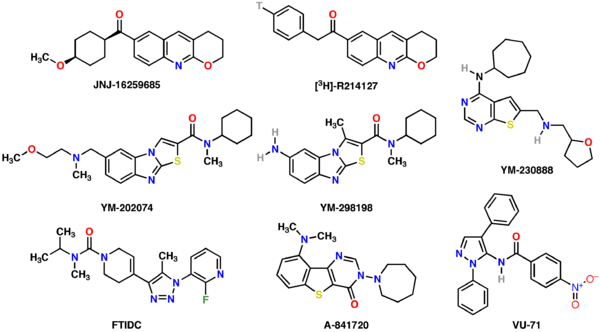
In addition to the orthosteric site (the site where the endogenous ligand glutamate binds) at least two distinct allosteric binding sites exist on the mGluR1.[11] A respectable number of potent and specific allosteric ligands – predominantly antagonists/inhibitors – has been developed in recent years, although no orthosteric subtype-selective ligands have yet been discovered (2008).[12]
- JNJ-16259685: highly potent, selective non-competitive antagonist[13]
- R-214,127 and [3H]-analog: high-affinity, selective allosteric inhibitor[14]
- YM-202,074: high-affinity, selective allosteric antagonist[15]
- YM-230,888: high-affinity, selective allosteric antagonist[16]
- YM-298,198 and [3H]-analog: selective non-competitive antagonist[17]
- FTIDC: highly potent and selective allosteric antagonist/inverse agonist[18]
- A-841,720: potent non-competitive antagonist; minor hmGluR5 binding[19]
- VU-71: potentiator[11]
- Fluorinated 9H-xanthene-9-carboxylic acid oxazol-2-yl-amides: orally available PAMs[20]
- Cyclothiazide: non-selective non-competitive antagonist[21]
See also
References
- GRCh38: Ensembl release 89: ENSG00000152822 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000019828 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: GRM1 glutamate receptor, metabotropic 1".
- Stephan D, Bon C, Holzwarth JA, Galvan M, Pruss RM (1996). "Human metabotropic glutamate receptor 1: mRNA distribution, chromosome localization and functional expression of two splice variants". Neuropharmacology. 35 (12): 1649–60. doi:10.1016/S0028-3908(96)00108-6. PMID 9076744.
- Makoff AJ, Phillips T, Pilling C, Emson P (September 1997). "Expression of a novel splice variant of human mGluR1 in the cerebellum". NeuroReport. 8 (13): 2943–7. doi:10.1097/00001756-199709080-00027. PMID 9376535.
- Smith RS, Weitz CJ, Araneda RC (Aug 2009). "Excitatory actions of noradrenaline and metabotropic glutamate receptor activation in granule cells of the accessory olfactory bulb". Journal of Neurophysiology. 102 (2): 1103–14. doi:10.1152/jn.91093.2008. PMC 2724365. PMID 19474170.
- Conquet F, Bashir ZI, Davies CH, Daniel H, Ferraguti F, Bordi F, Franz-Bacon K, Reggiani A, Matarese V, Condé F (November 1994). "Motor deficit and impairment of synaptic plasticity in mice lacking mGluR1". Nature. 372 (6503): 237–43. doi:10.1038/372237a0. PMID 7969468.
- Ortiz P, Vanaclocha F, López-Bran E, Esquivias JI, López-Estebaranz JL, Martín-González M, Arrue I, García-Romero D, Ochoa C, González-Perez A, Ruiz A, Real LM (November 2007). "Genetic analysis of the GRM1 gene in human melanoma susceptibility". Eur. J. Hum. Genet. 15 (11): 1176–82. doi:10.1038/sj.ejhg.5201887. PMID 17609672.
- Hemstapat K, de Paulis T, Chen Y, Brady AE, Grover VK, Alagille D, Tamagnan GD, Conn PJ (2006). "A novel class of positive allosteric modulators of metabotropic glutamate receptor subtype 1 interact with a site distinct from that of negative allosteric modulators". Mol. Pharmacol. 70 (2): 616–26. doi:10.1124/mol.105.021857. PMID 16645124.
- based on a plain pubmed review
- Lavreysen H, Wouters R, Bischoff F, Nóbrega Pereira S, Langlois X, Blokland S, Somers M, Dillen L, Lesage AS (2004). "JNJ16259685, a highly potent, selective and systemically active mGlu1 receptor antagonist". Neuropharmacology. 47 (7): 961–72. doi:10.1016/j.neuropharm.2004.08.007. PMID 15555631.
- Lavreysen H, Janssen C, Bischoff F, Langlois X, Leysen JE, Lesage AS (2003). "[3H]R214127: a novel high-affinity radioligand for the mGlu1 receptor reveals a common binding site shared by multiple allosteric antagonists". Mol. Pharmacol. 63 (5): 1082–93. doi:10.1124/mol.63.5.1082. PMID 12695537.
- Kohara A, Takahashi M, Yatsugi S, Tamura S, Shitaka Y, Hayashibe S, Kawabata S, Okada M (2008). "Neuroprotective effects of the selective type 1 metabotropic glutamate receptor antagonist YM-202074 in rat stroke models". Brain Res. 1191: 168–79. doi:10.1016/j.brainres.2007.11.035. PMID 18164695.
- Kohara A, Nagakura Y, Kiso T, Toya T, Watabiki T, Tamura S, Shitaka Y, Itahana H, Okada M (2007). "Antinociceptive profile of a selective metabotropic glutamate receptor 1 antagonist YM-230888 in chronic pain rodent models". Eur. J. Pharmacol. 571 (1): 8–16. doi:10.1016/j.ejphar.2007.05.030. PMID 17597604.
- Kohara A, Toya T, Tamura S, Watabiki T, Nagakura Y, Shitaka Y, Hayashibe S, Kawabata S, Okada M (2005). "Radioligand binding properties and pharmacological characterization of 6-amino-N-cyclohexyl-N,3-dimethylthiazolo[3,2-a]benzimidazole-2-carboxamide (YM-298198), a high-affinity, selective, and noncompetitive antagonist of metabotropic glutamate receptor type 1". J. Pharmacol. Exp. Ther. 315 (1): 163–9. doi:10.1124/jpet.105.087171. PMID 15976016.
- Suzuki G, Kimura T, Satow A, Kaneko N, Fukuda J, Hikichi H, Sakai N, Maehara S, Kawagoe-Takaki H, Hata M, Azuma T, Ito S, Kawamoto H, Ohta H (2007). "Pharmacological characterization of a new, orally active and potent allosteric metabotropic glutamate receptor 1 antagonist, 4-[1-(2-fluoropyridin-3-yl)-5-methyl-1H-1,2,3-triazol-4-yl]-N-isopropyl-N-methyl-3,6-dihydropyridine-1(2H)-carboxamide (FTIDC)". J. Pharmacol. Exp. Ther. 321 (3): 1144–53. doi:10.1124/jpet.106.116574. PMID 17360958.
- El-Kouhen O, Lehto SG, Pan JB, Chang R, Baker SJ, Zhong C, Hollingsworth PR, Mikusa JP, Cronin EA, Chu KL, McGaraughty SP, Uchic ME, Miller LN, Rodell NM, Patel M, Bhatia P, Mezler M, Kolasa T, Zheng GZ, Fox GB, Stewart AO, Decker MW, Moreland RB, Brioni JD, Honore P (2006). "Blockade of mGluR1 receptor results in analgesia and disruption of motor and cognitive performances: effects of A-841720, a novel non-competitive mGluR1 receptor antagonist". Br. J. Pharmacol. 149 (6): 761–74. doi:10.1038/sj.bjp.0706877. PMC 2014656. PMID 17016515.
- Vieira E, Huwyler J, Jolidon S, Knoflach F, Mutel V, Wichmann J (2009). "Fluorinated 9H-xanthene-9-carboxylic acid oxazol-2-yl-amides as potent, orally available mGlu1 receptor enhancers". Bioorg. Med. Chem. Lett. 19 (6): 1666–9. doi:10.1016/j.bmcl.2009.01.108. PMID 19233648.
- Surin A, Pshenichkin S, Grajkowska E, Surina E, Wroblewski JT (2007). "Cyclothiazide selectively inhibits mGluR1 receptors interacting with a common allosteric site for non-competitive antagonists". Neuropharmacology. 52 (3): 744–54. doi:10.1016/j.neuropharm.2006.09.018. PMC 1876747. PMID 17095021.
Further reading
- Bockaert J, Pin JP (1999). "Molecular tinkering of G protein-coupled receptors: an evolutionary success". EMBO J. 18 (7): 1723–9. doi:10.1093/emboj/18.7.1723. PMC 1171258. PMID 10202136.
- King JE, Eugenin EA, Buckner CM, Berman JW (2006). "HIV tat and neurotoxicity". Microbes Infect. 8 (5): 1347–57. doi:10.1016/j.micinf.2005.11.014. PMID 16697675.
- Desai MA, Burnett JP, Mayne NG, Schoepp DD (1995). "Cloning and expression of a human metabotropic glutamate receptor 1 alpha: enhanced coupling on co-transfection with a glutamate transporter". Mol. Pharmacol. 48 (4): 648–57. PMID 7476890.
- Scherer SW, Duvoisin RM, Kuhn R, Heng HH, Belloni E, Tsui LC (1997). "Localization of two metabotropic glutamate receptor genes, GRM3 and GRM8, to human chromosome 7q". Genomics. 31 (2): 230–3. doi:10.1006/geno.1996.0036. PMID 8824806.
- Brakeman PR, Lanahan AA, O'Brien R, Roche K, Barnes CA, Huganir RL, Worley PF (1997). "Homer: a protein that selectively binds metabotropic glutamate receptors". Nature. 386 (6622): 284–8. doi:10.1038/386284a0. PMID 9069287.
- Stephan D, Bon C, Holzwarth JA, Galvan M, Pruss RM (1997). "Human metabotropic glutamate receptor 1: mRNA distribution, chromosome localization and functional expression of two splice variants". Neuropharmacology. 35 (12): 1649–60. doi:10.1016/S0028-3908(96)00108-6. PMID 9076744.
- Makoff AJ, Phillips T, Pilling C, Emson P (1997). "Expression of a novel splice variant of human mGluR1 in the cerebellum". NeuroReport. 8 (13): 2943–7. doi:10.1097/00001756-199709080-00027. PMID 9376535.
- Francesconi A, Duvoisin RM (1998). "Role of the second and third intracellular loops of metabotropic glutamate receptors in mediating dual signal transduction activation". J. Biol. Chem. 273 (10): 5615–24. doi:10.1074/jbc.273.10.5615. PMID 9488690.
- Okamoto T, Sekiyama N, Otsu M, Shimada Y, Sato A, Nakanishi S, Jingami H (1998). "Expression and purification of the extracellular ligand binding region of metabotropic glutamate receptor subtype 1". J. Biol. Chem. 273 (21): 13089–96. doi:10.1074/jbc.273.21.13089. PMID 9582347.
- Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, Siderovski DP (1998). "GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain". J. Biol. Chem. 273 (28): 17749–55. doi:10.1074/jbc.273.28.17749. PMID 9651375.
- Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF (1998). "Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins". Neuron. 21 (4): 707–16. doi:10.1016/S0896-6273(00)80588-7. PMID 9808458.
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF (1998). "Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors". Neuron. 21 (4): 717–26. doi:10.1016/S0896-6273(00)80589-9. PMID 9808459.
- Ciruela F, Robbins MJ, Willis AC, McIlhinney RA (1999). "Interactions of the C terminus of metabotropic glutamate receptor type 1alpha with rat brain proteins: evidence for a direct interaction with tubulin". J. Neurochem. 72 (1): 346–54. doi:10.1046/j.1471-4159.1999.0720346.x. PMID 9886087.
- Robbins MJ, Ciruela F, Rhodes A, McIlhinney RA (1999). "Characterization of the dimerization of metabotropic glutamate receptors using an N-terminal truncation of mGluR1alpha". J. Neurochem. 72 (6): 2539–47. doi:10.1046/j.1471-4159.1999.0722539.x. PMID 10349865.
- Mody N, Hermans E, Nahorski SR, Challiss RA (1999). "Inhibition of N-linked glycosylation of the human type 1alpha metabotropic glutamate receptor by tunicamycin: effects on cell-surface receptor expression and function". Neuropharmacology. 38 (10): 1485–92. doi:10.1016/S0028-3908(99)00099-4. PMID 10530810.
- Francesconi A, Duvoisin RM (2000). "Opposing effects of protein kinase C and protein kinase A on metabotropic glutamate receptor signaling: selective desensitization of the inositol trisphosphate/Ca2+ pathway by phosphorylation of the receptor-G protein-coupling domain". Proc. Natl. Acad. Sci. U.S.A. 97 (11): 6185–90. doi:10.1073/pnas.97.11.6185. PMC 18579. PMID 10823959.
- Ganesh S, Amano K, Yamakawa K (2000). "Assignment of the gene GRM1 coding for metabotropic glutamate receptor 1 to human chromosome band 6q24 by in situ hybridization". Cytogenet. Cell Genet. 88 (3–4): 314–5. doi:10.1159/000015517. PMID 10828618.
- Ray K, Hauschild BC (2000). "Cys-140 is critical for metabotropic glutamate receptor-1 dimerization". J. Biol. Chem. 275 (44): 34245–51. doi:10.1074/jbc.M005581200. PMID 10945991.
- Hartley JL, Temple GF, Brasch MA (2001). "DNA cloning using in vitro site-specific recombination". Genome Res. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
External links
- "Metabotropic Glutamate Receptors: mGlu1". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology.
- Overview of all the structural information available in the PDB for UniProt: Q13255 (Metabotropic glutamate receptor 1) at the PDBe-KB.