Eukaryote
Eukaryotes (/juːˈkærioʊts, -əts/) are organisms whose cells have a nucleus enclosed within a nuclear envelope.[3][4][5] Eukaryotes belong to the domain Eukaryota or Eukarya; their name comes from the Greek εὖ (eu, "well" or "good") and κάρυον (karyon, "nut" or "kernel").[6] The domain Eukaryota makes up one of the domains of life in the three-domain system; the two other domains are Bacteria and Archaea (together known as prokaryotes).[7][8] Eukaryotes represent a tiny minority of the number of living organisms;[9] however, due to their generally much larger size, their collective worldwide biomass is estimated to be about equal to that of prokaryotes.[9] Eukaryotes evolved approximately 1.6–2.1 billion years ago, during the Proterozoic eon.
| Eukaryote | |
|---|---|
 | |
| Eukaryotes and some examples of their diversity – clockwise from top left: Red mason bee, Boletus edulis, chimpanzee, Isotricha intestinalis, Ranunculus asiaticus, and Volvox carteri | |
| Scientific classification | |
| Domain: | Eukaryota (Chatton, 1925) Whittaker & Margulis, 1978 |
| Supergroups[1] and kingdoms | |
Eukaryotic organisms that cannot be classified under the kingdoms Plantae, Animalia or Fungi are sometimes grouped in the kingdom Protista. | |
Eukaryotic cells typically contain membrane-bound organelles such as mitochondria and Golgi apparatus, and chloroplasts can be found in plants and algae; these organelles are unique to eukaryotes, although primitive organelles can be found in prokaryotes.[10] As well as being unicellular, eukaryotes may also be multicellular and include many cell types forming different kinds of tissue; in comparison, prokaryotes are typically unicellular. Animals, plants, and fungi are the most familiar eukaryotes; other eukaryotes are sometimes called protists.[11]
Eukaryotes can reproduce both asexually through mitosis and sexually through meiosis and gamete fusion. In mitosis, one cell divides to produce two genetically identical cells. In meiosis, DNA replication is followed by two rounds of cell division to produce four haploid daughter cells. These act as sex cells (gametes). Each gamete has just one set of chromosomes, each a unique mix of the corresponding pair of parental chromosomes resulting from genetic recombination during meiosis.[12]
History of the concept

The concept of the eukaryote has been attributed to the French biologist Edouard Chatton (1883–1947). The terms prokaryote and eukaryote were more definitively reintroduced by the Canadian microbiologist Roger Stanier and the Dutch-American microbiologist C. B. van Niel in 1962. In his 1937 work Titres et Travaux Scientifiques,[13] Chatton had proposed the two terms, calling the bacteria prokaryotes and organisms with nuclei in their cells eukaryotes. However he mentioned this in only one paragraph, and the idea was effectively ignored until Chatton's statement was rediscovered by Stanier and van Niel.[14]
In 1905 and 1910, the Russian biologist Konstantin Mereschkowski (1855–1921) argued that plastids were reduced cyanobacteria in a symbiosis with a non-photosynthetic (heterotrophic) host that was itself formed by symbiosis between an amoeba-like host and a bacterium-like cell that formed the nucleus. Plants had thus inherited photosynthesis from cyanobacteria.[15]
In 1967, Lynn Margulis provided microbiological evidence for endosymbiosis as the origin of chloroplasts and mitochondria in eukaryotic cells in her paper, On the origin of mitosing cells.[16] In the 1970s, Carl Woese explored microbial phylogenetics, studying variations in 16S ribosomal RNA. This helped to uncover the origin of the eukaryotes and the symbiogenesis of two important eukaryote organelles, mitochondria and chloroplasts. In 1977, Woese and George Fox introduced a "third form of life", which they called the Archaebacteria; in 1990, Woese, Otto Kandler and Mark L. Wheelis renamed this the Archaea.[17][14]
In 1979, G. W. Gould and G. J. Dring suggested that the eukaryotic cell's nucleus came from the ability of Gram-positive bacteria to form endospores. In 1987 and later papers, Thomas Cavalier-Smith proposed instead that the membranes of the nucleus and endoplasmic reticulum first formed by infolding a prokaryote's plasma membrane. In the 1990s, several other biologists proposed endosymbiotic origins for the nucleus, effectively reviving Mereschkowski's theory.[15]
Cell features
Eukaryotic cells are typically much larger than those of prokaryotes, having a volume of around 10,000 times greater than the prokaryotic cell.[18] They have a variety of internal membrane-bound structures, called organelles, and a cytoskeleton composed of microtubules, microfilaments, and intermediate filaments, which play an important role in defining the cell's organization and shape. Eukaryotic DNA is divided into several linear bundles called chromosomes, which are separated by a microtubular spindle during nuclear division.
Internal membrane
.svg.png)
Eukaryote cells include a variety of membrane-bound structures, collectively referred to as the endomembrane system.[19] Simple compartments, called vesicles and vacuoles, can form by budding off other membranes. Many cells ingest food and other materials through a process of endocytosis, where the outer membrane invaginates and then pinches off to form a vesicle.[20] It is probable that most other membrane-bound organelles are ultimately derived from such vesicles. Alternatively some products produced by the cell can leave in a vesicle through exocytosis.
The nucleus is surrounded by a double membrane (commonly referred to as a nuclear membrane or nuclear envelope), with pores that allow material to move in and out.[21] Various tube- and sheet-like extensions of the nuclear membrane form the endoplasmic reticulum, which is involved in protein transport and maturation. It includes the rough endoplasmic reticulum where ribosomes are attached to synthesize proteins, which enter the interior space or lumen. Subsequently, they generally enter vesicles, which bud off from the smooth endoplasmic reticulum.[22] In most eukaryotes, these protein-carrying vesicles are released and further modified in stacks of flattened vesicles (cisternae), the Golgi apparatus.[23]
Vesicles may be specialized for various purposes. For instance, lysosomes contain digestive enzymes that break down most biomolecules in the cytoplasm.[24] Peroxisomes are used to break down peroxide, which is otherwise toxic. Many protozoans have contractile vacuoles, which collect and expel excess water, and extrusomes, which expel material used to deflect predators or capture prey. In higher plants, most of a cell's volume is taken up by a central vacuole, which mostly contains water and primarily maintains its osmotic pressure.
Mitochondria and plastids
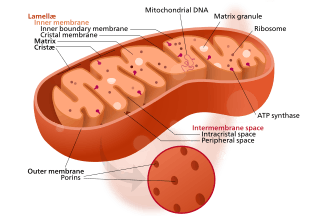
Mitochondria are organelles found in all but one[note 1] eukaryote. Mitochondria provide energy to the eukaryote cell by converting sugars into ATP.[26] They have two surrounding membranes, each a phospholipid bi-layer; the inner of which is folded into invaginations called cristae where aerobic respiration takes place.
The outer mitochondrial membrane is freely permeable and allows almost anything to enter into the intermembrane space while the inner mitochondrial membrane is semi permeable so allows only some required things into the mitochondrial matrix.
Mitochondria contain their own DNA, which has close structural similarities to bacterial DNA, and which encodes rRNA and tRNA genes that produce RNA which is closer in structure to bacterial RNA than to eukaryote RNA.[27] They are now generally held to have developed from endosymbiotic prokaryotes, probably proteobacteria.
Some eukaryotes, such as the metamonads such as Giardia and Trichomonas, and the amoebozoan Pelomyxa, appear to lack mitochondria, but all have been found to contain mitochondrion-derived organelles, such as hydrogenosomes and mitosomes, and thus have lost their mitochondria secondarily.[25] They obtain energy by enzymatic action on nutrients absorbed from the environment. The metamonad Monocercomonoides has also acquired, by lateral gene transfer, a cytosolic sulfur mobilisation system which provides the clusters of iron and sulfur required for protein synthesis. The normal mitochondrial iron-sulfur cluster pathway has been lost secondarily.[25][28]
Plants and various groups of algae also have plastids. Plastids also have their own DNA and are developed from endosymbionts, in this case cyanobacteria. They usually take the form of chloroplasts which, like cyanobacteria, contain chlorophyll and produce organic compounds (such as glucose) through photosynthesis. Others are involved in storing food. Although plastids probably had a single origin, not all plastid-containing groups are closely related. Instead, some eukaryotes have obtained them from others through secondary endosymbiosis or ingestion.[29] The capture and sequestering of photosynthetic cells and chloroplasts occurs in many types of modern eukaryotic organisms and is known as kleptoplasty.
Endosymbiotic origins have also been proposed for the nucleus, and for eukaryotic flagella.[30]
Cytoskeletal structures
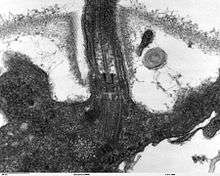
Many eukaryotes have long slender motile cytoplasmic projections, called flagella, or similar structures called cilia. Flagella and cilia are sometimes referred to as undulipodia,[31] and are variously involved in movement, feeding, and sensation. They are composed mainly of tubulin. These are entirely distinct from prokaryotic flagellae. They are supported by a bundle of microtubules arising from a centriole, characteristically arranged as nine doublets surrounding two singlets. Flagella also may have hairs, or mastigonemes, and scales connecting membranes and internal rods. Their interior is continuous with the cell's cytoplasm.
Microfilamental structures composed of actin and actin binding proteins, e.g., α-actinin, fimbrin, filamin are present in submembranous cortical layers and bundles, as well. Motor proteins of microtubules, e.g., dynein or kinesin and actin, e.g., myosins provide dynamic character of the network.
Centrioles are often present even in cells and groups that do not have flagella, but conifers and flowering plants have neither. They generally occur in groups that give rise to various microtubular roots. These form a primary component of the cytoskeletal structure, and are often assembled over the course of several cell divisions, with one flagellum retained from the parent and the other derived from it. Centrioles produce the spindle during nuclear division.[32]
The significance of cytoskeletal structures is underlined in the determination of shape of the cells, as well as their being essential components of migratory responses like chemotaxis and chemokinesis. Some protists have various other microtubule-supported organelles. These include the radiolaria and heliozoa, which produce axopodia used in flotation or to capture prey, and the haptophytes, which have a peculiar flagellum-like organelle called the haptonema.
Cell wall
The cells of plants and algae, fungi and most chromalveolates have a cell wall, a layer outside the cell membrane, providing the cell with structural support, protection, and a filtering mechanism. The cell wall also prevents over-expansion when water enters the cell.[33]
The major polysaccharides making up the primary cell wall of land plants are cellulose, hemicellulose, and pectin. The cellulose microfibrils are linked via hemicellulosic tethers to form the cellulose-hemicellulose network, which is embedded in the pectin matrix. The most common hemicellulose in the primary cell wall is xyloglucan.[34]
Differences among eukaryotic cells
There are many different types of eukaryotic cells, though animals and plants are the most familiar eukaryotes, and thus provide an excellent starting point for understanding eukaryotic structure. Fungi and many protists have some substantial differences, however.
Animal cell

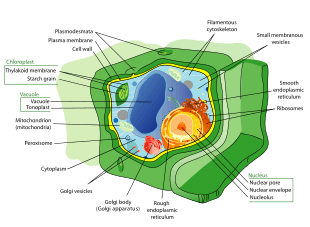
All animals are eukaryotic. Animal cells are distinct from those of other eukaryotes, most notably plants, as they lack cell walls and chloroplasts and have smaller vacuoles. Due to the lack of a cell wall, animal cells can transform into a variety of shapes. A phagocytic cell can even engulf other structures.
Plant cell
Plant cells are quite different from the cells of the other eukaryotic organisms. Their distinctive features are:
- A large central vacuole (enclosed by a membrane, the tonoplast), which maintains the cell's turgor and controls movement of molecules between the cytosol and sap[35]
- A primary cell wall containing cellulose, hemicellulose and pectin, deposited by the protoplast on the outside of the cell membrane; this contrasts with the cell walls of fungi, which contain chitin, and the cell envelopes of prokaryotes, in which peptidoglycans are the main structural molecules
- The plasmodesmata, pores in the cell wall that link adjacent cells and allow plant cells to communicate with adjacent cells.[36] Animals have a different but functionally analogous system of gap junctions between adjacent cells.
- Plastids, especially chloroplasts, organelles that contain chlorophyll, the pigment that gives plants their green color and allows them to perform photosynthesis
- Bryophytes and seedless vascular plants only have flagellae and centrioles in the sperm cells.[37] Sperm of cycads and Ginkgo are large, complex cells that swim with hundreds to thousands of flagellae.[38]
- Conifers (Pinophyta) and flowering plants (Angiospermae) lack the flagellae and centrioles that are present in animal cells.
Fungal cell
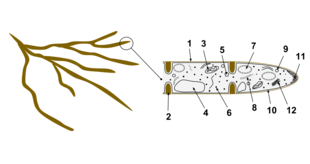
The cells of fungi are most similar to animal cells, with the following exceptions:[39]
- A cell wall that contains chitin
- Less compartmentation between cells; the hyphae of higher fungi have porous partitions called septa, which allow the passage of cytoplasm, organelles, and, sometimes, nuclei; so each organism is essentially a giant multinucleate supercell — these fungi are described as coenocytic. Primitive fungi have few or no septa.
- Only the most primitive fungi, chytrids, have flagella.
Other eukaryotic cells
Some groups of eukaryotes have unique organelles, such as the cyanelles (unusual chloroplasts) of the glaucophytes,[40] the haptonema of the haptophytes, or the ejectosomes of the cryptomonads. Other structures, such as pseudopodia, are found in various eukaryote groups in different forms, such as the lobose amoebozoans or the reticulose foraminiferans.[41]
Reproduction
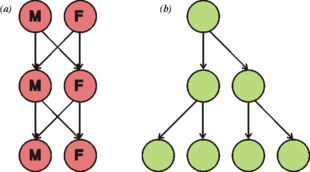
Cell division generally takes place asexually by mitosis, a process that allows each daughter nucleus to receive one copy of each chromosome. Most eukaryotes also have a life cycle that involves sexual reproduction, alternating between a haploid phase, where only one copy of each chromosome is present in each cell and a diploid phase, wherein two copies of each chromosome are present in each cell. The diploid phase is formed by fusion of two haploid gametes to form a zygote, which may divide by mitosis or undergo chromosome reduction by meiosis. There is considerable variation in this pattern. Animals have no multicellular haploid phase, but each plant generation can consist of haploid and diploid multicellular phases.
Eukaryotes have a smaller surface area to volume ratio than prokaryotes, and thus have lower metabolic rates and longer generation times.[42]
The evolution of sexual reproduction may be a primordial and fundamental characteristic of eukaryotes. Based on a phylogenetic analysis, Dacks and Roger proposed that facultative sex was present in the common ancestor of all eukaryotes.[43] A core set of genes that function in meiosis is present in both Trichomonas vaginalis and Giardia intestinalis, two organisms previously thought to be asexual.[44][45] Since these two species are descendants of lineages that diverged early from the eukaryotic evolutionary tree, it was inferred that core meiotic genes, and hence sex, were likely present in a common ancestor of all eukaryotes.[44][45] Eukaryotic species once thought to be asexual, such as parasitic protozoa of the genus Leishmania, have been shown to have a sexual cycle.[46] Also, evidence now indicates that amoebae, previously regarded as asexual, are anciently sexual and that the majority of present-day asexual groups likely arose recently and independently.[47]
Classification
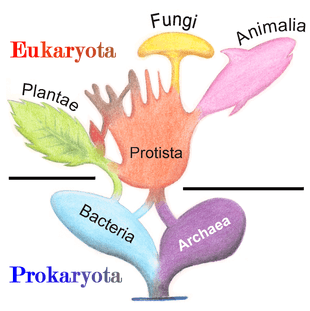
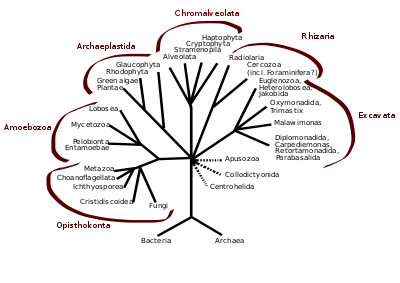
In antiquity, the two lineages of animals and plants were recognized. They were given the taxonomic rank of Kingdom by Linnaeus. Though he included the fungi with plants with some reservations, it was later realized that they are quite distinct and warrant a separate kingdom, the composition of which was not entirely clear until the 1980s.[48] The various single-cell eukaryotes were originally placed with plants or animals when they became known. In 1818, the German biologist Georg A. Goldfuss coined the word protozoa to refer to organisms such as ciliates,[49] and this group was expanded until it encompassed all single-celled eukaryotes, and given their own kingdom, the Protista, by Ernst Haeckel in 1866.[50][51] The eukaryotes thus came to be composed of four kingdoms:
The protists were understood to be "primitive forms", and thus an evolutionary grade, united by their primitive unicellular nature.[51] The disentanglement of the deep splits in the tree of life only really started with DNA sequencing, leading to a system of domains rather than kingdoms as top level rank being put forward by Carl Woese, uniting all the eukaryote kingdoms under the eukaryote domain.[17] At the same time, work on the protist tree intensified, and is still actively going on today. Several alternative classifications have been forwarded, though there is no consensus in the field.
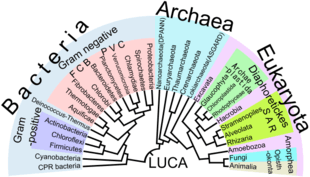
Eukaryotes are a clade usually assessed to be sister to Heimdallarchaeota in the Asgard grouping in the Archaea.[52][53][54] The basal groupings are the Opimoda, Diphoda, the Discoba, and the Loukozoa. The Eukaryote root is usually assessed to be near or even in Discoba.
A classification produced in 2005 for the International Society of Protistologists,[55] which reflected the consensus of the time, divided the eukaryotes into six supposedly monophyletic 'supergroups'. However, in the same year (2005), doubts were expressed as to whether some of these supergroups were monophyletic, particularly the Chromalveolata,[56] and a review in 2006 noted the lack of evidence for several of the supposed six supergroups.[57] A revised classification in 2012[1] recognizes five supergroups.
| Archaeplastida (or Primoplantae) | Land plants, green algae, red algae, and glaucophytes |
| SAR supergroup | Stramenopiles (brown algae, diatoms, etc.), Alveolata, and Rhizaria (Foraminifera, Radiolaria, and various other amoeboid protozoa) |
| Excavata | Various flagellate protozoa |
| Amoebozoa | Most lobose amoeboids and slime molds |
| Opisthokonta | Animals, fungi, choanoflagellates, etc. |
There are also smaller groups of eukaryotes whose position is uncertain or seems to fall outside the major groups[58] – in particular, Haptophyta, Cryptophyta, Centrohelida, Telonemia, Picozoa,[59] Apusomonadida, Ancyromonadida, Breviatea, and the genus Collodictyon.[60] Overall, it seems that, although progress has been made, there are still very significant uncertainties in the evolutionary history and classification of eukaryotes. As Roger & Simpson said in 2009 "with the current pace of change in our understanding of the eukaryote tree of life, we should proceed with caution."[61]
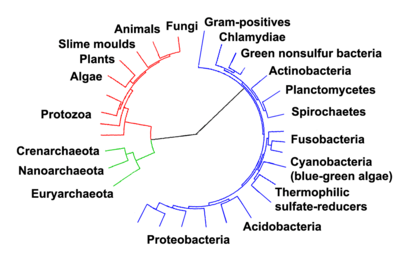
In an article published in Nature Microbiology in April 2016 the authors, "reinforced once again that the life we see around us – plants, animals, humans and other so-called eukaryotes – represent a tiny percentage of the world's biodiversity."[62] They classified eukaryote "based on the inheritance of their information systems as opposed to lipid or other cellular structures." Jillian F. Banfield of the University of California, Berkeley and fellow scientists used a super computer to generate a diagram of a new tree of life based on DNA from 3000 species including 2,072 known species and 1,011 newly reported microbial organisms, whose DNA they had gathered from diverse environments.[8][63] As the capacity to sequence DNA became easier, Banfield and team were able to do metagenomic sequencing – "sequencing whole communities of organisms at once and picking out the individual groups based on their genes alone."[62]
Phylogeny
The rRNA trees constructed during the 1980s and 1990s left most eukaryotes in an unresolved "crown" group (not technically a true crown), which was usually divided by the form of the mitochondrial cristae; see crown eukaryotes. The few groups that lack mitochondria branched separately, and so the absence was believed to be primitive; but this is now considered an artifact of long-branch attraction, and they are known to have lost them secondarily.[64][65]
As of 2011, there is widespread agreement that the Rhizaria belong with the Stramenopiles and the Alveolata, in a clade dubbed the SAR supergroup, so that Rhizaria is not one of the main eukaryote groups; also that the Amoebozoa and Opisthokonta are each monophyletic and form a clade, often called the unikonts.[66][67][68][69][70] Beyond this, there does not appear to be a consensus.
It has been estimated that there may be 75 distinct lineages of eukaryotes.[71] Most of these lineages are protists.
The known eukaryote genome sizes vary from 8.2 megabases (Mb) in Babesia bovis to 112,000–220,050 Mb in the dinoflagellate Prorocentrum micans, showing that the genome of the ancestral eukaryote has undergone considerable variation during its evolution.[71] The last common ancestor of all eukaryotes is believed to have been a phagotrophic protist with a nucleus, at least one centriole and cilium, facultatively aerobic mitochondria, sex (meiosis and syngamy), a dormant cyst with a cell wall of chitin and/or cellulose and peroxisomes.[71] Later endosymbiosis led to the spread of plastids in some lineages.
Five supergroups
A global tree of eukaryotes from a consensus of phylogenetic evidence (in particular, phylogenomics), rare genomic signatures, and morphological characteristics is presented in Adl et al. 2012[1] and Burki 2014/2016 with the Cryptophyta and picozoa having emerged within the Archaeplastida.[58][72][73][74][75][76] A similar inclusion of Glaucophyta, Cryptista (and also, unusually, Haptista) has also been made.[25]
| Eukaryotes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
In some analyses, the Hacrobia group (Haptophyta + Cryptophyta) is placed next to Archaeplastida,[66] but in other ones it is nested inside the Archaeplastida.[77] However, several recent studies have concluded that Haptophyta and Cryptophyta do not form a monophyletic group.[78] The former could be a sister group to the SAR group, the latter cluster with the Archaeplastida (plants in the broad sense).[79]
The division of the eukaryotes into two primary clades, bikonts (Archaeplastida + SAR + Excavata) and unikonts (Amoebozoa + Opisthokonta), derived from an ancestral biflagellar organism and an ancestral uniflagellar organism, respectively, had been suggested earlier.[77][80][81] A 2012 study produced a somewhat similar division, although noting that the terms "unikonts" and "bikonts" were not used in the original sense.[59]
A highly converged and congruent set of trees appears in Derelle et al. (2015), Ren et al. (2016), Yang et al. (2017) and Cavalier-Smith (2015) including the supplementary information, resulting in a more conservative and consolidated tree. It is combined with some results from Cavalier-Smith for the basal Opimoda.[82][83][84][85][86][75][87] The main remaining controversies are the root, and the exact positioning of the Rhodophyta and the bikonts Rhizaria, Haptista, Cryptista, Picozoa and Telonemia, many of which may be endosymbiotic eukaryote-eukaryote hybrids.[88] Archaeplastida acquired chloroplasts probably by endosymbiosis of a prokaryotic ancestor related to a currently extant cyanobacterium, Gloeomargarita lithophora.[89][90][88]
| Eukaryotes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Cavalier-Smith's tree
Thomas Cavalier-Smith 2010,[91] 2013,[92] 2014,[93] 2017[83] and 2018[94] places the eukaryotic tree's root between Excavata (with ventral feeding groove supported by a microtubular root) and the grooveless Euglenozoa, and monophyletic Chromista, correlated to a single endosymbiotic event of capturing a red-algae. He et al.[95] specifically supports rooting the eukaryotic tree between a monophyletic Discoba (Discicristata + Jakobida) and an Amorphea-Diaphoretickes clade.
| Eukaryotes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
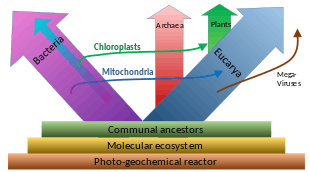
Origin of eukaryotes
The origin of the eukaryotic cell is a milestone in the evolution of life, since eukaryotes include all complex cells and almost all multicellular organisms. A number of approaches have been used to find the first eukaryote and their closest relatives. The last eukaryotic common ancestor (LECA) is the hypothetical last common ancestor of all eukaryotes that have ever lived, and was most likely a biological population.[98]
Eukaryotes have a set of signature features that differentiate them from other domains of life, including an endomembrane system and unique biochemical pathways such as sterane synthesis.[99] A set of proteins called eukaryotic signature proteins (ESPs) was proposed to identify eukaryotic relatives in 2002: they have no homology to proteins known in other domains of life by then, but they appear to be universal among eukaryotes. They include proteins that make up the cytoskeleton, the complex transcription machinery, membrane-sorting systems, the nuclear pore, as well as some enzymes in the biochemical pathways.[100]
Fossils
The timing of this series of events is hard to determine; Knoll (2006) suggests they developed approximately 1.6–2.1 billion years ago. Some acritarchs are known from at least 1.65 billion years ago, and the possible alga Grypania has been found as far back as 2.1 billion years ago.[101] The Geosiphon-like fossil fungus Diskagma has been found in paleosols 2.2 billion years old.[102]
Organized living structures have been found in the black shales of the Palaeoproterozoic Francevillian B Formation in Gabon, dated at 2.1 billion years old. Eukaryotic life could have evolved at that time.[103] Fossils that are clearly related to modern groups start appearing an estimated 1.2 billion years ago, in the form of a red algae, though recent work suggests the existence of fossilized filamentous algae in the Vindhya basin dating back perhaps to 1.6 to 1.7 billion years ago.[104]
Biomarkers suggest that at least stem eukaryotes arose even earlier. The presence of steranes in Australian shales indicates that eukaryotes were present in these rocks dated at 2.7 billion years old,[99][105] although it was suggested they could originate from samples contamination.[106]
Whenever their origins, eukaryotes may not have become ecologically dominant until much later; a massive uptick in the zinc composition of marine sediments 800 million years ago has been attributed to the rise of substantial populations of eukaryotes, which preferentially consume and incorporate zinc relative to prokaryotes.[107]
In April 2019, biologists reported that the very large medusavirus, or a relative, may have been responsible, at least in part, for the evolutionary emergence of complex eukaryotic cells from simpler prokaryotic cells.[108]
Relationship to Archaea
The nuclear DNA and genetic machinery of eukaryotes is more similar to Archaea than Bacteria, leading to a controversial suggestion that eukaryotes should be grouped with Archaea in the clade Neomura. In other respects, such as membrane composition, eukaryotes are similar to Bacteria. Three main explanations for this have been proposed:
- Eukaryotes resulted from the complete fusion of two or more cells, wherein the cytoplasm formed from a eubacterium, and the nucleus from an archaeon,[109] from a virus,[110][111] or from a pre-cell.[112][113]
- Eukaryotes developed from Archaea, and acquired their eubacterial characteristics through the endosymbiosis of a proto-mitochondrion of eubacterial origin.[114]
- Eukaryotes and Archaea developed separately from a modified eubacterium.
Alternative proposals include:
- The chronocyte hypothesis postulates that a primitive eukaryotic cell was formed by the endosymbiosis of both archaea and bacteria by a third type of cell, termed a chronocyte. This is mainly to account for the fact that eukaryotic signature proteins were not found anywhere else by 2002.[100]
- The universal common ancestor (UCA) of the current tree of life was a complex organism that survived a mass extinction event rather than an early stage in the evolution of life. Eukaryotes and in particular akaryotes (Bacteria and Archaea) evolved through reductive loss, so that similarities result from differential retention of original features.[116]
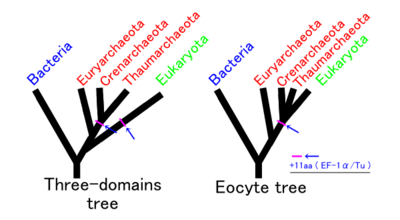
Assuming no other group is involved, there are three possible phylogenies for the Bacteria, Archaea and Eukaryota in which each is monophyletic. These are labelled 1 to 3 in the table below. The eocyte hypothesis is a modification of hypothesis 2 in which the Archaea are paraphyletic. (The table and the names for the hypotheses are based on Harish and Kurland, 2017.[117])
| 1 – Two empires | 2 – Three domains | 3 – Gupta | 4 – Eocyte | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
In recent years, most researchers have favoured either the three domains (3D) or the eocyte hypothesis. An rRNA analyses supports the eocyte scenario, apparently with the Eukaryote root in Excavata.[63][91][92][93][83] A cladogram supporting the eocyte hypothesis, positioning eukaryotes within Archaea, based on phylogenomic analyses of the Asgard archaea, is:[52][53][54]
| Proteoarchaeota |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
In this scenario, the Asgard group is seen as a sister taxon of the TACK group, which comprises Crenarchaeota (formerly named eocytes), Thaumarchaeota, and others. This group is reported contain many of the eukaryotic signature proteins and produce vesicles.[118]
In 2017, there has been significant pushback against this scenario, arguing that the eukaryotes did not emerge within the Archaea. Cunha et al. produced analyses supporting the three domains (3D) or Woese hypothesis (2 in the table above) and rejecting the eocyte hypothesis (4 above).[119] Harish and Kurland found strong support for the earlier two empires (2D) or Mayr hypothesis (1 in the table above), based on analyses of the coding sequences of protein domains. They rejected the eocyte hypothesis as the least likely.[120][117] A possible interpretation of their analysis is that the universal common ancestor (UCA) of the current tree of life was a complex organism that survived an evolutionary bottleneck, rather than a simpler organism arising early in the history of life.[116] On the other hand, the researchers who came up with Asgard re-affirmed their hypothesis with additional Asgard samples.[121]
Details of the relation of Asgard archaea members and eukaryotes are still under consideration,[122] although, in January 2020, scientists reported that Candidatus Prometheoarchaeum syntrophicum, a type of cultured Asgard archaea, may be a possible link between simple prokaryotic and complex eukaryotic microorganisms about two billion years ago.[123][118]
Endomembrane system and mitochondria
The origins of the endomembrane system and mitochondria are also unclear.[124] The phagotrophic hypothesis proposes that eukaryotic-type membranes lacking a cell wall originated first, with the development of endocytosis, whereas mitochondria were acquired by ingestion as endosymbionts.[125] The syntrophic hypothesis proposes that the proto-eukaryote relied on the proto-mitochondrion for food, and so ultimately grew to surround it. Here the membranes originated after the engulfment of the mitochondrion, in part thanks to mitochondrial genes (the hydrogen hypothesis is one particular version).[126]
In a study using genomes to construct supertrees, Pisani et al. (2007) suggest that, along with evidence that there was never a mitochondrion-less eukaryote, eukaryotes evolved from a syntrophy between an archaea closely related to Thermoplasmatales and an α-proteobacterium, likely a symbiosis driven by sulfur or hydrogen. The mitochondrion and its genome is a remnant of the α-proteobacterial endosymbiont.[127] The majority of the genes from the symbiont have been transferred to the nucleus. They make up most of the metabolic and energy-related pathways of the eukaryotic cell, while the information system is retained from archaea.[128]
Hypotheses
Different hypotheses have been proposed as to how eukaryotic cells came into existence. These hypotheses can be classified into two distinct classes – autogenous models and chimeric models.
Autogenous models
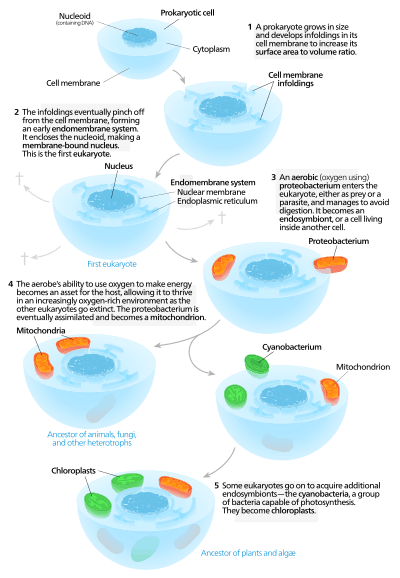
Autogenous models propose that a proto-eukaryotic cell containing a nucleus existed first, and later acquired mitochondria.[129] According to this model, a large prokaryote developed invaginations in its plasma membrane in order to obtain enough surface area to service its cytoplasmic volume. As the invaginations differentiated in function, some became separate compartments – giving rise to the endomembrane system, including the endoplasmic reticulum, golgi apparatus, nuclear membrane, and single membrane structures such as lysosomes.[130]
Mitochondria are proposed to come from the endosymbiosis of an aerobic proteobacterium, and it is assumed that all the eukaryotic lineages that did not acquire mitochondria became extinct,[131] a statement criticized for its lack of falsifiability. Chloroplasts came about from another endosymbiotic event involving cyanobacteria. Since all known eukaryotes have mitochondria, but not all have chloroplasts, the serial endosymbiosis theory proposes that mitochondria came first.
Chimeric models
Chimeric models claim that two prokaryotic cells existed initially – an archaeon and a bacterium. The closest living relatives of these appears to be Asgardarchaeota and (distantly related) the alphaproteobacteria.[132][133] These cells underwent a merging process, either by a physical fusion or by endosymbiosis, thereby leading to the formation of a eukaryotic cell. Within these chimeric models, some studies further claim that mitochondria originated from a bacterial ancestor while others emphasize the role of endosymbiotic processes behind the origin of mitochondria.
The inside-out hypothesis
The inside-out hypothesis, developed by cousins David and Buzz Baum, suggest the fusion between free-living mitochondria-like bacteria and an archaeon into a eukaryotic cell happened gradually over a long period of time, instead of phagocytosis in a single gulp. In this scenario, an archaeon would trap aerobic bacteria with cell protrusions, and then keep them alive to draw energy from them instead of digesting them. During the early stages the bacteria would still be partly in direct contact with the environment, and the archaeon would not have to provide them with all the required nutrients. But eventually the archaeon would engulf the bacteria completely, creating the internal membrane structures and nucleus membrane in the process.[134]
It is assumed the archaean group called halophiles went through a similar procedure, where they acquired as much as a thousand genes from a bacterium, way more than through the conventional horizontal gene transfer that often occurs in the microbial world, but that the two microbes separated again before they had fused into a single eukaryote-like cell.[135]
Based on the process of mutualistic symbiosis, the hypotheses can be categorized as – the serial endosymbiotic hypothesis or theory (SET),[136][137][138] the hydrogen hypothesis (mostly a process of symbiosis where hydrogen transfer takes place among different species),[126] and the syntrophy hypothesis.[139][140] These hypotheses are discussed separately in the following sections.
An expanded version of the inside-out hypothesis proposes that the eukaryotic cell was created by physical interactions between two prokaryotic organisms and that the last common ancestor of eukaryotes got its genome from a whole population or community of microbes participating in cooperative relationships to thrive and survive in their environment. The genome from the various types of microbes would complement each other, and occasional horizontal gene transfer between them would be largely to their own benefit. This accumulation of beneficial genes gave rise to the genome of the eukaryotic cell, which contained all the genes required for independence.[141]
The serial endosymbiotic hypothesis
According to serial endosymbiotic theory (championed by Lynn Margulis), a union between a motile anaerobic bacterium (like Spirochaeta) and a thermoacidophilic crenarchaeon (like Thermoplasma which is sulfidogenic in nature) gave rise to the present day eukaryotes. This union established a motile organism capable of living in the already existing acidic and sulfurous waters. Oxygen is known to cause toxicity to organisms that lack the required metabolic machinery. Thus, the archaeon provided the bacterium with a highly beneficial reduced environment (sulfur and sulfate were reduced to sulfide). In microaerophilic conditions, oxygen was reduced to water thereby creating a mutual benefit platform. The bacterium on the other hand, contributed the necessary fermentation products and electron acceptors along with its motility feature to the archaeon thereby gaining a swimming motility for the organism.
From a consortium of bacterial and archaeal DNA originated the nuclear genome of eukaryotic cells. Spirochetes gave rise to the motile features of eukaryotic cells. Endosymbiotic unifications of the ancestors of alpha-proteobacteria and cyanobacteria, led to the origin of mitochondria and plastids respectively. For example, Thiodendron has been known to have originated via an ectosymbiotic process based on a similar syntrophy of sulfur existing between the two types of bacteria – Desulphobacter and Spirochaeta.
However, such an association based on motile symbiosis has never been observed practically. Also there is no evidence of archaeans and spirochetes adapting to intense acid-based environments.[129]
The hydrogen hypothesis
In the hydrogen hypothesis, the symbiotic linkage of an anaerobic and autotrophic methanogenic archaeon (host) with an alpha-proteobacterium (the symbiont) gave rise to the eukaryotes. The host utilized hydrogen (H2) and carbon dioxide (CO
2) to produce methane while the symbiont, capable of aerobic respiration, expelled H2 and CO
2 as byproducts of anaerobic fermentation process. The host's methanogenic environment worked as a sink for H2, which resulted in heightened bacterial fermentation.
Endosymbiotic gene transfer (EGT) acted as a catalyst for the host to acquire the symbionts' carbohydrate metabolism and turn heterotrophic in nature. Subsequently, the host's methane forming capability was lost. Thus, the origins of the heterotrophic organelle (symbiont) are identical to the origins of the eukaryotic lineage. In this hypothesis, the presence of H2 represents the selective force that forged eukaryotes out of prokaryotes.
The syntrophy hypothesis
The syntrophy hypothesis was developed in contrast to the hydrogen hypothesis and proposes the existence of two symbiotic events. According to this theory, the origin of eukaryotic cells was based on metabolic symbiosis (syntrophy) between a methanogenic archaeon and a delta-proteobacterium. This syntrophic symbiosis was initially facilitated by H2 transfer between different species under anaerobic environments. In earlier stages, an alpha-proteobacterium became a member of this integration, and later developed into the mitochondrion. Gene transfer from a delta-proteobacterium to an archaeon led to the methanogenic archaeon developing into a nucleus. The archaeon constituted the genetic apparatus, while the delta-proteobacterium contributed towards the cytoplasmic features.
This theory incorporates two selective forces at the time of nucleus evolution
- presence of metabolic partitioning to avoid the harmful effects of the co-existence of anabolic and catabolic cellular pathways, and
- prevention of abnormal protein biosynthesis due to a vast spread of introns in the archaeal genes after acquiring the mitochondrion and losing methanogenesis.
6+ serial endosymbiosis scenario
Pitts and Galbanón propose a complex scenario of 6+ serial endosymbiotic events of Archaea and bacteria in which mitochondria and an asgard related archaeota were acquired at a late stage of eukaryogenesis, possibly in combination, as a secondary endosymbiont.[142][143] The findings have been rebuked as an artefact.[144]
See also
Notes
- To date, only one eukaryote, Monocercomonoides, is known to have completely lost its mitochondria.[25]
References
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, et al. (September 2012). "The revised classification of eukaryotes" (PDF). The Journal of Eukaryotic Microbiology. 59 (5): 429–93. doi:10.1111/j.1550-7408.2012.00644.x. PMC 3483872. PMID 23020233. Archived from the original (PDF) on 16 June 2016.
- Sakaguchi M, Takishita K, Matsumoto T, Hashimoto T, Inagaki Y (July 2009). "Tracing back EFL gene evolution in the cryptomonads-haptophytes assemblage: separate origins of EFL genes in haptophytes, photosynthetic cryptomonads, and goniomonads". Gene. 441 (1–2): 126–31. doi:10.1016/j.gene.2008.05.010. PMID 18585873.
- Youngson RM (2006). Collins Dictionary of Human Biology. Glasgow: HarperCollins. ISBN 978-0-00-722134-9.
- Nelson DL, Cox MM (2005). Lehninger Principles of Biochemistry (4th ed.). New York: W.H. Freeman. ISBN 978-0-7167-4339-2.
- Martin EA, ed. (1983). Macmillan Dictionary of Life Sciences (2nd ed.). London: Macmillan Press. ISBN 978-0-333-34867-3.
- Harper, Douglas. "eukaryotic". Online Etymology Dictionary.
- Woese CR, Kandler O, Wheelis ML (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–9. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Zimmer C (11 April 2016). "Scientists Unveil New 'Tree of Life'". The New York Times. Retrieved 11 April 2016.
- Whitman WB, Coleman DC, Wiebe WJ (June 1998). "Prokaryotes: the unseen majority" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 95 (12): 6578–6583. Bibcode:1998PNAS...95.6578W. doi:10.1073/pnas.95.12.6578. PMC 33863. PMID 9618454.
- Murat, Dorothee; Byrne, Meghan; Komeili, Arash (1 October 2010). "Cell Biology of Prokaryotic Organelles". Cold Spring Harbor Perspectives in Biology. doi:10.1101/cshperspect.a000422. PMC 2944366. PMID 20739411. Retrieved 11 July 2020.
- Whittaker, R.H. (January 1969). "New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms". Science. 163 (3863): 150–60. Bibcode:1969Sci...163..150W. CiteSeerX 10.1.1.403.5430. doi:10.1126/science.163.3863.150. PMID 5762760.
- Campbell NA, Cain ML, Minorsky PV, Reece JB, Urry LA (2018). "Chapter 13: Sexual Life Cycles and Meiosis". Biology: A Global Approach (11th ed.). New York: Pearson Education. ISBN 978-1-292-17043-5.
- Chatton, Édouard (1937). Titres Et Travaux Scientifiques (1906-1937) De Edouard Chatton. Sète: Impr. E. Sottano.
- Sapp J (June 2005). "The prokaryote-eukaryote dichotomy: meanings and mythology". Microbiology and Molecular Biology Reviews. 69 (2): 292–305. doi:10.1128/MMBR.69.2.292-305.2005. PMC 1197417. PMID 15944457.
- Martin WF, Garg S, Zimorski V (September 2015). "Endosymbiotic theories for eukaryote origin". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 370 (1678): 20140330. doi:10.1098/rstb.2014.0330. PMC 4571569. PMID 26323761.
- Sagan L (March 1967). "On the origin of mitosing cells". Journal of Theoretical Biology. 14 (3): 255–274. doi:10.1016/0022-5193(67)90079-3. PMID 11541392.
- Woese CR, Kandler O, Wheelis ML (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Yamaguchi M, Worman CO (2014). "Deep-sea microorganisms and the origin of the eukaryotic cell" (PDF). Jpn. J. Protozool. 47 (1, 2): 29–48. Archived from the original (PDF) on 9 August 2017. Retrieved 24 October 2017.
- Linka M, Weber AP (2011). "Evolutionary Integration of Chloroplast Metabolism with the Metabolic Networks of the Cells". In Burnap RL, Vermaas WF (eds.). Functional Genomics and Evolution of Photosynthetic Systems. Springer. p. 215. ISBN 978-9400715332.
- Marsh M (2001). Endocytosis. Oxford University Press. p. vii. ISBN 978-0-19-963851-2.
- Hetzer MW (March 2010). "The nuclear envelope". Cold Spring Harbor Perspectives in Biology. 2 (3): a000539. doi:10.1101/cshperspect.a000539. PMC 2829960. PMID 20300205.
- "Endoplasmic Reticulum (Rough and Smooth)". British Society for Cell Biology. Retrieved 12 November 2017.
- "Golgi Apparatus". British Society for Cell Biology. Archived from the original on 13 November 2017. Retrieved 12 November 2017.
- "Lysosome". British Society for Cell Biology. Archived from the original on 13 November 2017. Retrieved 12 November 2017.
- Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, Soukal P, Hroudová M, Doležal P, Stairs CW, Roger AJ, Eliáš M, Dacks JB, Vlček Č, Hampl V (May 2016). "A Eukaryote without a Mitochondrial Organelle". Current Biology. 26 (10): 1274–1284. doi:10.1016/j.cub.2016.03.053. PMID 27185558.
- Mack, Steve (1 May 2006). "Re: Are there eukaryotic cells without mitochondria?". madsci.org.
- Watson J, Hopkins N, Roberts J, Steitz JA, Weiner A (1988). "28: The Origins of Life". Molecular Biology of the Gene (Fourth ed.). Menlo Park, CA: The Benjamin/Cummings Publishing Company, Inc. p. 1154. ISBN 978-0-8053-9614-0.
- Davis JL (13 May 2016). "Scientists Shocked To Discover Eukaryote With NO Mitochondria". IFL Science. Archived from the original on 17 February 2019. Retrieved 13 May 2016.
- Sato N (2006). "Origin and Evolution of Plastids: Genomic View on the Unification and Diversity of Plastids". In Wise RR, Hoober JK (eds.). The Structure and Function of Plastids. Advances in Photosynthesis and Respiration. 23. Springer Netherlands. pp. 75–102. doi:10.1007/978-1-4020-4061-0_4. ISBN 978-1-4020-4060-3.
- Margulis L (1998). Symbiotic planet: a new look at evolution. New York: Basic Books. ISBN 978-0-465-07271-2. OCLC 39700477.
- Lynn Margulis, Heather I. McKhann & Lorraine Olendzenski (ed.), Illustrated Glossary of Protoctista, Jones and Bartlett Publishers, Boston, 1993, p. xviii. ISBN 0-86720-081-2
- Vorobjev IA, Nadezhdina ES (1987). The centrosome and its role in the organization of microtubules. International Review of Cytology. 106. pp. 227–293. doi:10.1016/S0074-7696(08)61714-3. ISBN 978-0-12-364506-7. PMID 3294718.
- Howland JL (2000). The Surprising Archaea: Discovering Another Domain of Life. Oxford: Oxford University Press. pp. 69–71. ISBN 978-0-19-511183-5.
- Fry, Stephen C. (1989). "The Structure and Functions of Xyloglucan". Journal of Experimental Botany. 40 (1): 1–11. doi:10.1093/jxb/40.1.1.
- Raven J (July 1987). "The role of vacuoles". New Phytologist. 106 (3): 357–422. doi:10.1111/j.1469-8137.1987.tb00149.x.
- Oparka K (2005). Plasmodesmata. Oxford, UK: Blackwell Publishing.
- Raven PH, Evert RF, Eichorm SE (1999). Biology of Plants. New York: W.H. Freeman.
- Silflow CD, Lefebvre PA (December 2001). "Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii". Plant Physiology. 127 (4): 1500–1507. doi:10.1104/pp.010807. PMC 1540183. PMID 11743094.
- Deacon J (2005). Fungal Biology. Cambridge, Massachusetts: Blackwell Publishers. pp. 4 and passim. ISBN 978-1-4051-3066-0.
- Keeling PJ (October 2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany. 91 (10): 1481–1493. doi:10.3732/ajb.91.10.1481. PMID 21652304.
- Patterson DJ. "Amoebae: Protists Which Move and Feed Using Pseudopodia". Tree of Life Web Project. Retrieved 12 November 2017.
- Lane N (June 2011). "Energetics and genetics across the prokaryote-eukaryote divide". Biology Direct. 6 (1): 35. doi:10.1186/1745-6150-6-35. PMC 3152533. PMID 21714941.
- Dacks J, Roger AJ (June 1999). "The first sexual lineage and the relevance of facultative sex". Journal of Molecular Evolution. 48 (6): 779–783. Bibcode:1999JMolE..48..779D. doi:10.1007/PL00013156. PMID 10229582. S2CID 9441768.
- Ramesh MA, Malik SB, Logsdon JM (January 2005). "A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis". Current Biology. 15 (2): 185–191. doi:10.1016/j.cub.2005.01.003. PMID 15668177.
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM (August 2007). Hahn MW (ed.). "An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis". PLOS ONE. 3 (8): e2879. Bibcode:2008PLoSO...3.2879M. doi:10.1371/journal.pone.0002879. PMC 2488364. PMID 18663385.
- Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, Dobson DE, Beverley SM, Sacks DL (April 2009). "Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector". Science. 324 (5924): 265–268. Bibcode:2009Sci...324..265A. doi:10.1126/science.1169464. PMC 2729066. PMID 19359589.
- Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E (July 2011). "The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms". Proceedings: Biological Sciences. 278 (1715): 2081–2090. doi:10.1098/rspb.2011.0289. PMC 3107637. PMID 21429931.
- Moore RT (1980). "Taxonomic proposals for the classification of marine yeasts and other yeast-like fungi including the smuts". Botanica Marina. 23: 361–373.
- Goldfuß (1818). "Ueber die Classification der Zoophyten" [On the classification of zoophytes]. Isis, Oder, Encyclopädische Zeitung von Oken (in German). 2 (6): 1008–1019. From p. 1008: "Erste Klasse. Urthiere. Protozoa." (First class. Primordial animals. Protozoa.) [Note: each column of each page of this journal is numbered; there are two columns per page.]
- Scamardella JM (1999). "Not plants or animals: a brief history of the origin of Kingdoms Protozoa, Protista and Protoctista" (PDF). International Microbiology. 2: 207–221. PMID 10943416. Archived from the original (PDF) on 14 June 2011.
- Rothschild LJ (1989). "Protozoa, Protista, Protoctista: what's in a name?". Journal of the History of Biology. 22 (2): 277–305. doi:10.1007/BF00139515. PMID 11542176.
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ (May 2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature. 521 (7551): 173–179. Bibcode:2015Natur.521..173S. doi:10.1038/nature14447. PMC 4444528. PMID 25945739.
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJ (January 2017). "Asgard archaea illuminate the origin of eukaryotic cellular complexity". Nature. 541 (7637): 353–358. Bibcode:2017Natur.541..353Z. doi:10.1038/nature21031. PMID 28077874.
- Liu Y, Zhou Z, Pan J, Baker BJ, Gu JD, Li M (April 2018). "Comparative genomic inference suggests mixotrophic lifestyle for Thorarchaeota". The ISME Journal. 12 (4): 1021–1031. doi:10.1038/s41396-018-0060-x. PMC 5864231. PMID 29445130.
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. (2005). "The new higher level classification of eukaryotes with emphasis on the taxonomy of protists". The Journal of Eukaryotic Microbiology. 52 (5): 399–451. doi:10.1111/j.1550-7408.2005.00053.x. PMID 16248873.
- Harper JT, Waanders E, Keeling PJ (January 2005). "On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes" (PDF). International Journal of Systematic and Evolutionary Microbiology. 55 (Pt 1): 487–496. doi:10.1099/ijs.0.63216-0. PMID 15653923. Archived from the original (PDF) on 17 December 2008.
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA (December 2006). "Evaluating support for the current classification of eukaryotic diversity". PLOS Genetics. 2 (12): e220. doi:10.1371/journal.pgen.0020220. PMC 1713255. PMID 17194223.
- Burki F (May 2014). "The eukaryotic tree of life from a global phylogenomic perspective". Cold Spring Harbor Perspectives in Biology. 6 (5): a016147. doi:10.1101/cshperspect.a016147. PMC 3996474. PMID 24789819.
- Zhao S, Burki F, Bråte J, Keeling PJ, Klaveness D, Shalchian-Tabrizi K (June 2012). "Collodictyon – an ancient lineage in the tree of eukaryotes". Molecular Biology and Evolution. 29 (6): 1557–1568. doi:10.1093/molbev/mss001. PMC 3351787. PMID 22319147.
- Romari K, Vaulot D (2004). "Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences". Limnol Oceanogr. 49 (3): 784–798. Bibcode:2004LimOc..49..784R. doi:10.4319/lo.2004.49.3.0784. S2CID 86718111.
- Roger AJ, Simpson AG (February 2009). "Evolution: revisiting the root of the eukaryote tree". Current Biology. 19 (4): R165–67. doi:10.1016/j.cub.2008.12.032. PMID 19243692.
- Sanders R (11 April 2016). "Wealth of unsuspected new microbes expands tree of life". Berkeley News. Archived from the original on 20 April 2016. Retrieved 11 April 2016.
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF (April 2016). "A new view of the tree of life". Nature Microbiology. 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647.
- Tovar J, Fischer A, Clark CG (June 1999). "The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica". Molecular Microbiology. 32 (5): 1013–1021. doi:10.1046/j.1365-2958.1999.01414.x. PMID 10361303.
- Boxma B, de Graaf RM, van der Staay GW, van Alen TA, Ricard G, Gabaldón T, van Hoek AH, Moon-van der Staay SY, Koopman WJ, van Hellemond JJ, Tielens AG, Friedrich T, Veenhuis M, Huynen MA, Hackstein JH (March 2005). "An anaerobic mitochondrion that produces hydrogen" (PDF). Nature. 434 (7029): 74–79. Bibcode:2005Natur.434...74B. doi:10.1038/nature03343. PMID 15744302.
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J (August 2007). Butler G (ed.). "Phylogenomics reshuffles the eukaryotic supergroups". PLOS ONE. 2 (8): e790. Bibcode:2007PLoSO...2..790B. doi:10.1371/journal.pone.0000790. PMC 1949142. PMID 17726520.
- Burki F, Shalchian-Tabrizi K, Pawlowski J (August 2008). "Phylogenomics reveals a new 'megagroup' including most photosynthetic eukaryotes". Biology Letters. 4 (4): 366–369. doi:10.1098/rsbl.2008.0224. PMC 2610160. PMID 18522922.
- Burki F, Inagaki Y, Bråte J, Archibald JM, Keeling PJ, Cavalier-Smith T, Sakaguchi M, Hashimoto T, Horak A, Kumar S, Klaveness D, Jakobsen KS, Pawlowski J, Shalchian-Tabrizi K (July 2009). "Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, telonemia and centroheliozoa, are related to photosynthetic chromalveolates". Genome Biology and Evolution. 1: 231–238. doi:10.1093/gbe/evp022. PMC 2817417. PMID 20333193.
- Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rümmele SE, Bhattacharya D (August 2007). "Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates". Molecular Biology and Evolution. 24 (8): 1702–1713. doi:10.1093/molbev/msm089. PMID 17488740.
- Cavalier-Smith T (June 2010). "Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree". Biology Letters. 6 (3): 342–345. doi:10.1098/rsbl.2009.0948. PMC 2880060. PMID 20031978.
- Jagus R, Bachvaroff TR, Joshi B, Place AR (2012). "Diversity of Eukaryotic Translational Initiation Factor eIF4E in Protists". Comparative and Functional Genomics. 2012: 1–21. doi:10.1155/2012/134839. PMC 3388326. PMID 22778692.
- Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Smirnov A, Mylnikov AP, Keeling PJ (January 2016). "Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista". Proceedings: Biological Sciences. 283 (1823): 20152802. doi:10.1098/rspb.2015.2802. PMC 4795036. PMID 26817772.
- Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Rohwer FL, Mylnikov AP, Keeling PJ (December 2017). "A New Lineage of Eukaryotes Illuminates Early Mitochondrial Genome Reduction" (PDF). Current Biology. 27 (23): 3717–24.e5. doi:10.1016/j.cub.2017.10.051. PMID 29174886.
- Bodył A (February 2018). "Did some red alga-derived plastids evolve via kleptoplastidy? A hypothesis". Biological Reviews of the Cambridge Philosophical Society. 93 (1): 201–222. doi:10.1111/brv.12340. PMID 28544184. S2CID 24613863.
- Brown MW, Heiss AA, Kamikawa R, Inagaki Y, Yabuki A, Tice AK, Shiratori T, Ishida KI, Hashimoto T, Simpson AG, Roger AJ (February 2018). "Phylogenomics Places Orphan Protistan Lineages in a Novel Eukaryotic Super-Group". Genome Biology and Evolution. 10 (2): 427–433. doi:10.1093/gbe/evy014. PMC 5793813. PMID 29360967.
- Lax G, Eglit Y, Eme L, Bertrand EM, Roger AJ, Simpson AG (November 2018). "Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes". Nature. 564 (7736): 410–414. Bibcode:2018Natur.564..410L. doi:10.1038/s41586-018-0708-8. PMID 30429611. S2CID 205570993.
- Kim E, Graham LE (July 2008). Redfield RJ (ed.). "EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata". PLOS ONE. 3 (7): e2621. Bibcode:2008PLoSO...3.2621K. doi:10.1371/journal.pone.0002621. PMC 2440802. PMID 18612431.
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H (July 2010). "Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles". Molecular Biology and Evolution. 27 (7): 1698–1709. doi:10.1093/molbev/msq059. PMID 20194427.
- Burki F, Okamoto N, Pombert JF, Keeling PJ (June 2012). "The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins". Proceedings: Biological Sciences. 279 (1736): 2246–2254. doi:10.1098/rspb.2011.2301. PMC 3321700. PMID 22298847.
- Cavalier-Smith T (2006). "Protist phylogeny and the high-level classification of Protozoa". European Journal of Protistology. 39 (4): 338–348. doi:10.1078/0932-4739-00002. S2CID 84403388.
- Burki F, Pawlowski J (October 2006). "Monophyly of Rhizaria and multigene phylogeny of unicellular bikonts". Molecular Biology and Evolution. 23 (10): 1922–1930. doi:10.1093/molbev/msl055. PMID 16829542.
- Ren R, Sun Y, Zhao Y, Geiser D, Ma H, Zhou X (September 2016). "Phylogenetic Resolution of Deep Eukaryotic and Fungal Relationships Using Highly Conserved Low-Copy Nuclear Genes". Genome Biology and Evolution. 8 (9): 2683–2701. doi:10.1093/gbe/evw196. PMC 5631032. PMID 27604879.
- Cavalier-Smith T (January 2018). "Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences". Protoplasma. 255 (1): 297–357. doi:10.1007/s00709-017-1147-3. PMC 5756292. PMID 28875267.
- Derelle R, Torruella G, Klimeš V, Brinkmann H, Kim E, Vlček Č, Lang BF, Eliáš M (February 2015). "Bacterial proteins pinpoint a single eukaryotic root". Proceedings of the National Academy of Sciences of the United States of America. 112 (7): E693–699. Bibcode:2015PNAS..112E.693D. doi:10.1073/pnas.1420657112. PMC 4343179. PMID 25646484.
- Yang J, Harding T, Kamikawa R, Simpson AG, Roger AJ (May 2017). "Mitochondrial Genome Evolution and a Novel RNA Editing System in Deep-Branching Heteroloboseids". Genome Biology and Evolution. 9 (5): 1161–1174. doi:10.1093/gbe/evx086. PMC 5421314. PMID 28453770.
- Cavalier-Smith T, Fiore-Donno AM, Chao E, Kudryavtsev A, Berney C, Snell EA, Lewis R (February 2015). "Multigene phylogeny resolves deep branching of Amoebozoa". Molecular Phylogenetics and Evolution. 83: 293–304. doi:10.1016/j.ympev.2014.08.011. PMID 25150787.
- Torruella G, de Mendoza A, Grau-Bové X, Antó M, Chaplin MA, del Campo J, Eme L, Pérez-Cordón G, Whipps CM, Nichols KM, Paley R, Roger AJ, Sitjà-Bobadilla A, Donachie S, Ruiz-Trillo I (September 2015). "Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi". Current Biology. 25 (18): 2404–2410. doi:10.1016/j.cub.2015.07.053. PMID 26365255.
- López-García P, Eme L, Moreira D (December 2017). "Symbiosis in eukaryotic evolution". Journal of Theoretical Biology. 434: 20–33. doi:10.1016/j.jtbi.2017.02.031. PMC 5638015. PMID 28254477.
- Ponce-Toledo RI, Deschamps P, López-García P, Zivanovic Y, Benzerara K, Moreira D (February 2017). "An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids". Current Biology. 27 (3): 386–391. doi:10.1016/j.cub.2016.11.056. PMC 5650054. PMID 28132810.
- de Vries J, Archibald JM (February 2017). "Endosymbiosis: Did Plastids Evolve from a Freshwater Cyanobacterium?". Current Biology. 27 (3): R103–105. doi:10.1016/j.cub.2016.12.006. PMID 28171752.
- Cavalier-Smith T (June 2010). "Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree". Biology Letters. 6 (3): 342–345. doi:10.1098/rsbl.2009.0948. PMC 2880060. PMID 20031978.
- Cavalier-Smith T (May 2013). "Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa". European Journal of Protistology. 49 (2): 115–178. doi:10.1016/j.ejop.2012.06.001. PMID 23085100.
- Cavalier-Smith T, Chao EE, Snell EA, Berney C, Fiore-Donno AM, Lewis R (December 2014). "Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa". Molecular Phylogenetics and Evolution. 81: 71–85. doi:10.1016/j.ympev.2014.08.012. PMID 25152275.
- Cavalier-Smith T, Chao EE, Lewis R (April 2018). "Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria". Protoplasma. 255 (5): 1517–1574. doi:10.1007/s00709-018-1241-1. PMC 6133090. PMID 29666938.
- He D, Fiz-Palacios O, Fu CJ, Fehling J, Tsai CC, Baldauf SL (February 2014). "An alternative root for the eukaryote tree of life". Current Biology. 24 (4): 465–470. doi:10.1016/j.cub.2014.01.036. PMID 24508168.
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM (December 2008). "The archaebacterial origin of eukaryotes". Proceedings of the National Academy of Sciences of the United States of America. 105 (51): 20356–20361. Bibcode:2008PNAS..10520356C. doi:10.1073/pnas.0810647105. PMC 2629343. PMID 19073919.
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (March 2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–1287. Bibcode:2006Sci...311.1283C. CiteSeerX 10.1.1.381.9514. doi:10.1126/science.1123061. PMID 16513982.
- O’Malley, Maureen A.; Leger, Michelle M.; Wideman, Jeremy G.; Ruiz-Trillo, Iñaki (18 February 2019). "Concepts of the last eukaryotic common ancestor". Nature Ecology & Evolution. Springer Science and Business Media LLC. 3 (3): 338–344. doi:10.1038/s41559-019-0796-3. ISSN 2397-334X.
- Brocks JJ, Logan GA, Buick R, Summons RE (August 1999). "Archean molecular fossils and the early rise of eukaryotes". Science. 285 (5430): 1033–1036. CiteSeerX 10.1.1.516.9123. doi:10.1126/science.285.5430.1033. PMID 10446042.
- Hartman H, Fedorov A (February 2002). "The origin of the eukaryotic cell: a genomic investigation". Proceedings of the National Academy of Sciences of the United States of America. 99 (3): 1420–5. Bibcode:2002PNAS...99.1420H. doi:10.1073/pnas.032658599. PMC 122206. PMID 11805300.
- Knoll AH, Javaux EJ, Hewitt D, Cohen P (June 2006). "Eukaryotic organisms in Proterozoic oceans". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 361 (1470): 1023–1038. doi:10.1098/rstb.2006.1843. PMC 1578724. PMID 16754612.
- Retallack GJ, Krull ES, Thackray GD, Parkinson DH (2013). "Problematic urn-shaped fossils from a Paleoproterozoic (2.2 Ga) paleosol in South Africa". Precambrian Research. 235: 71–87. Bibcode:2013PreR..235...71R. doi:10.1016/j.precamres.2013.05.015.
- El Albani A, Bengtson S, Canfield DE, Bekker A, Macchiarelli R, Mazurier A, Hammarlund EU, Boulvais P, Dupuy JJ, Fontaine C, Fürsich FT, Gauthier-Lafaye F, Janvier P, Javaux E, Ossa FO, Pierson-Wickmann AC, Riboulleau A, Sardini P, Vachard D, Whitehouse M, Meunier A (July 2010). "Large colonial organisms with coordinated growth in oxygenated environments 2.1 Gyr ago". Nature. 466 (7302): 100–104. Bibcode:2010Natur.466..100A. doi:10.1038/nature09166. PMID 20596019. S2CID 4331375.
- Bengtson S, Belivanova V, Rasmussen B, Whitehouse M (May 2009). "The controversial "Cambrian" fossils of the Vindhyan are real but more than a billion years older". Proceedings of the National Academy of Sciences of the United States of America. 106 (19): 7729–7734. Bibcode:2009PNAS..106.7729B. doi:10.1073/pnas.0812460106. PMC 2683128. PMID 19416859.
- Ward P (9 February 2008). "Mass extinctions: the microbes strike back". New Scientist: 40–43.
- French KL, Hallmann C, Hope JM, Schoon PL, Zumberge JA, Hoshino Y, Peters CA, George SC, Love GD, Brocks JJ, Buick R, Summons RE (May 2015). "Reappraisal of hydrocarbon biomarkers in Archean rocks". Proceedings of the National Academy of Sciences of the United States of America. 112 (19): 5915–5920. Bibcode:2015PNAS..112.5915F. doi:10.1073/pnas.1419563112. PMC 4434754. PMID 25918387.
- Isson TT, Love GD, Dupont CL, Reinhard CT, Zumberge AJ, Asael D, et al. (June 2018). "Tracking the rise of eukaryotes to ecological dominance with zinc isotopes". Geobiology. 16 (4): 341–352. doi:10.1111/gbi.12289. PMID 29869832.
- Yoshikawa G, Blanc-Mathieu R, Song C, Kayama Y, Mochizuki T, Murata K, Ogata H, Takemura M (April 2019). "Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water". Journal of Virology. 93 (8). doi:10.1128/JVI.02130-18. PMC 6450098. PMID 30728258. Lay summary – EurekAlert! (30 April 2019).
- Martin W (December 2005). "Archaebacteria (Archaea) and the origin of the eukaryotic nucleus". Current Opinion in Microbiology. 8 (6): 630–637. doi:10.1016/j.mib.2005.10.004. PMID 16242992.
- Takemura M (May 2001). "Poxviruses and the origin of the eukaryotic nucleus". Journal of Molecular Evolution. 52 (5): 419–425. Bibcode:2001JMolE..52..419T. doi:10.1007/s002390010171. PMID 11443345. S2CID 21200827.
- Bell PJ (September 2001). "Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus?". Journal of Molecular Evolution. 53 (3): 251–256. Bibcode:2001JMolE..53..251L. doi:10.1007/s002390010215. PMID 11523012. S2CID 20542871.
- Wächtershäuser G (January 2003). "From pre-cells to Eukarya – a tale of two lipids". Molecular Microbiology. 47 (1): 13–22. doi:10.1046/j.1365-2958.2003.03267.x. PMID 12492850. S2CID 37944519.
- Wächtershäuser G (October 2006). "From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 361 (1474): 1787–1806, discussion 1806–1808. doi:10.1098/rstb.2006.1904. PMC 1664677. PMID 17008219.
- Lane, Nick (2016). The Vital Question: Why is Life the Way it is? (paperback ed.). Profile Books. pp. 157–91. ISBN 978-1-781-25037-2.
- Egel R (January 2012). "Primal eukaryogenesis: on the communal nature of precellular States, ancestral to modern life". Life. 2 (1): 170–212. doi:10.3390/life2010170. PMC 4187143. PMID 25382122.
- Harish A, Tunlid A, Kurland CG (August 2013). "Rooted phylogeny of the three superkingdoms". Biochimie. 95 (8): 1593–1604. doi:10.1016/j.biochi.2013.04.016. PMID 23669449.
- Harish A, Kurland CG (July 2017). "Akaryotes and Eukaryotes are independent descendants of a universal common ancestor". Biochimie. 138: 168–183. doi:10.1016/j.biochi.2017.04.013. PMID 28461155.
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. (January 2020). "Isolation of an archaeon at the prokaryote-eukaryote interface". Nature. 577 (7791): 519–525. doi:10.1038/s41586-019-1916-6. PMC 7015854. PMID 31942073.
- Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P (June 2017). "Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes". PLOS Genetics. 13 (6): e1006810. doi:10.1371/journal.pgen.1006810. PMC 5484517. PMID 28604769.
- Harish A, Kurland CG (July 2017). "Empirical genome evolution models root the tree of life". Biochimie. 138: 137–155. doi:10.1016/j.biochi.2017.04.014. PMID 28478110.
- Spang A, Eme L, Saw JH, Caceres EF, Zaremba-Niedzwiedzka K, Lombard J, et al. (March 2018). "Asgard archaea are the closest prokaryotic relatives of eukaryotes". PLOS Genetics. 14 (3): e1007080. doi:10.1371/journal.pgen.1007080. PMC 5875740. PMID 29596421.
- MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP (2019). "Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes". AIMS Microbiology. 5 (1): 48–61. doi:10.3934/microbiol.2019.1.48. PMC 6646929. PMID 31384702.
- Zimmer, Carl (15 January 2020). "This Strange Microbe May Mark One of Life's Great Leaps - A organism living in ocean muck offers clues to the origins of the complex cells of all animals and plants". The New York Times. Retrieved 18 January 2020.
- Jékely G (2007). "Origin of Eukaryotic Endomembranes: A Critical Evaluation of Different Model Scenarios". Eukaryotic Membranes and Cytoskeleton. Advances in Experimental Medicine and Biology. 607. New York, N.Y. : Springer Science+Business Media; Austin, Tex. : Landes Bioscience. pp. 38–51. doi:10.1007/978-0-387-74021-8_3. ISBN 978-0-387-74020-1. PMID 17977457.
- Cavalier-Smith T (March 2002). "The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa". International Journal of Systematic and Evolutionary Microbiology. 52 (Pt 2): 297–354. doi:10.1099/00207713-52-2-297. PMID 11931142.
- Martin W, Müller M (March 1998). "The hydrogen hypothesis for the first eukaryote". Nature. 392 (6671): 37–41. Bibcode:1998Natur.392...37M. doi:10.1038/32096. PMID 9510246. S2CID 338885.
- Pisani D, Cotton JA, McInerney JO (August 2007). "Supertrees disentangle the chimerical origin of eukaryotic genomes". Molecular Biology and Evolution. 24 (8): 1752–1760. doi:10.1093/molbev/msm095. PMID 17504772.
- Brueckner J, Martin WF (April 2020). "Bacterial Genes Outnumber Archaeal Genes in Eukaryotic Genomes". Genome Biology and Evolution. 12 (4): 282–292. doi:10.1093/gbe/evaa047. PMC 7151554. PMID 32142116.
- Latorre A, Durban A, Moya A, Pereto J (2011). "The role of symbiosis in eukaryotic evolution". In Gargaud M, López-Garcìa P, Martin H (eds.). Origins and Evolution of Life: An astrobiological perspective. Cambridge: Cambridge University Press. pp. 326–339. ISBN 978-0-521-76131-4.
- Ayala J (April 1994). "Transport and internal organization of membranes: vesicles, membrane networks and GTP-binding proteins". Journal of Cell Science. 107 ( Pt 4) (107): 753–763. PMID 8056835. Archived from the original on 29 April 2012. Retrieved 27 March 2013.
- Martin WF. "The Origin of Mitochondria". Scitable. Nature education. Retrieved 27 March 2013.
- Dacks JB, Field MC (August 2018). "Evolutionary origins and specialisation of membrane transport". Current Opinion in Cell Biology. 53: 70–76. doi:10.1016/j.ceb.2018.06.001. PMC 6141808. PMID 29929066.
- Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJ (May 2018). "Deep mitochondrial origin outside the sampled alphaproteobacteria". Nature. 557 (7703): 101–105. Bibcode:2018Natur.557..101M. doi:10.1038/s41586-018-0059-5. PMID 29695865. S2CID 13740626. Lay summary – The Scientist.
- Baum DA, Baum B (October 2014). "An inside-out origin for the eukaryotic cell". BMC Biology. 12: 76. doi:10.1186/s12915-014-0076-2. PMC 4210606. PMID 25350791. Lay summary – University of Wisconsin-Madison.
- Brouwers L (12 April 2013). "How genetic plunder transformed a microbe into a pink, salt-loving scavenger". Scientific American. 109 (50): 20537–20542. Archived from the original on 10 October 2018. Retrieved 21 April 2019.
- Margulis L (1970). Origin of Eukaryotic Cells. New Haven, London: Yale University Press.
- Margulis L (1993). Symbiosis in Cell Evolution. New York: W.H. Freeman.
- Margulis L, Dolan MF, Guerrero R (June 2000). "The chimeric eukaryote: Origin of the nucleus from the karyomastigont in amitochondriate protists". Proceedings of the National Academy of Sciences of the United States of America. 97 (13): 6954–6959. Bibcode:2000PNAS...97.6954M. doi:10.1073/pnas.97.13.6954. PMC 34369. PMID 10860956.
- Moreira D, Lopez-Garcia P (November 1998). "Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: The syntrophic hypothesis". Journal of Molecular Evolution. 47 (5): 517–530. Bibcode:1998JMolE..47..517M. doi:10.1007/PL00006408. PMID 9797402. S2CID 3911443.
- López-García P, Moreira D (May 2006). "Selective forces for the origin of the eukaryotic nucleus". BioEssays. 28 (5): 525–533. doi:10.1002/bies.20413. PMID 16615090.
- "Rethinking the ancestry of the eukaryotes". Quanta Magazine. Archived from the original on 9 May 2019. Retrieved 8 May 2019.
- Pittis AA, Gabaldón T (March 2016). "Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry". Nature. 531 (7592): 101–104. Bibcode:2016Natur.531..101P. doi:10.1038/nature16941. PMC 4780264. PMID 26840490.
- Burton ZF (1 August 2017). Evolution since coding: Cradles, halos, barrels, and wings. Academic Press. ISBN 9780128130346.
- Martin WF, Roettger M, Ku C, Garg SG, Nelson-Sathi S, Landan G (February 2017). "Late mitochondrial origin is an artifact". Genome Biology and Evolution. 9 (2): 373–379. doi:10.1093/gbe/evx027. PMC 5516564. PMID 28199635.
![]()
.jpg)
_(cropped).jpg)