Archamoebae
The Archamoebae are a group of protists originally thought to have evolved before the acquisition of mitochondria by eukaryotes.[1] They include genera that are internal parasites or commensals of animals (Entamoeba and Endolimax). A few species are human pathogens, causing diseases such as amoebic dysentery. The other genera of archamoebae live in freshwater habitats and are unusual among amoebae in possessing flagella. Most have a single nucleus and flagellum, but the giant amoeba Pelomyxa has many of each.
| Archamoebae | |
|---|---|
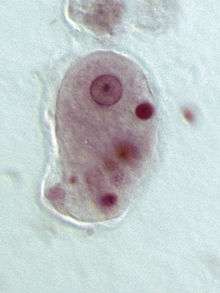 | |
| Entamoeba histolytica trophozoite | |
| Scientific classification | |
| Domain: | Eukaryota |
| (unranked): | Unikonta |
| Phylum: | Amoebozoa |
| Subphylum: | Conosa |
| Infraphylum: | Archamoebae Cavalier-Smith 1998 |
| Families and orders | |
| |
| Synonyms | |
| |
Description
Archamoebae are a diverse group of amoebae. Many have flagella for motility, while others do not. They grow in the absence of oxygen, though some can tolerate small amounts. Most described species of Archamoebae either lack mitochondria or are described to have reduced mitosomes.[2]
Classification
History
The group Archamoebae was proposed by Thomas Cavalier-Smith in 1998 as part of the Archezoa, a newly-proposed group to include eukaryotes that had diverged before acquisition of mitochondria and other common eukaryotic cell features.[3][4] Early molecular trees based on rRNA supported this position, placing several Archamoebae genera as separate groups that diverged from other eukaryotes very early on, suggesting that the absence of mitochondria was a primitive condition.[4] However, soon thereafter genetic remnants of mitochondria were found in various Archamoebae, suggesting that these organisms had diverged after the evolution of mitochondria, but had lost their mitochondria over time, and are more closely related to various amoebae and slime molds.[1]
Taxonomy
Infraphylum Archamoebae Cavalier-Smith 1993 stat. nov. 1998[5][6][7][8]
- Class Archamoebea Cavalier-Smith 1983 stat. nov. 2004
- Family Tricholimacidae Cavalier-Smith 2013
- Genus Tricholimax Frenzel 1892
- Family Endamoebidae alkins 1926
- Genus Endamoeba Leidy 1879
- Order Entamoebida Cavalier-Smith 1993
- Family Entamoebidae Chatton 1925 em. Cavalier-Smith 1993
- Genus †Entamoebites Poinar & Boucot 2006
- Genus Entamoeba Casagrandi & Barbagallo 1895
- Family Entamoebidae Chatton 1925 em. Cavalier-Smith 1993
- Order Pelobiontida Page 1976 em. Cavalier Smith 1987
- Suborder Pelomyxina Starobogatov 1980
- Family Pelomyxidae Shulze 1877 em. Cavalier-Smith 2016
- Genus Pelomyxa Greeff 1874
- Genus Mastigella Frenzel 1892
- Family Pelomyxidae Shulze 1877 em. Cavalier-Smith 2016
- Suborder Mastigamoebina (Frenzel 1897) Pánek et al. 2016
- Family Rhizomastigidae Cavalier-Smith 2013
- Genus Rhizomastix Aléxéieff 1911
- Family Mastigamoebidae Goldschmidt 1907
- ?Genus Craigia Calkins 1913
- ?Genus Dobellina Bishop & Tate 1940
- ?Genus Pansporella Chatton 1925
- ?Genus Martineziella Hegner & Hewitt 1941 non Chalumeau 1986
- ?Genus Dinamoeba Leidy 1874 non Pascher 1916
- Genus Endolimax Kuenen & Swellengrebel 1917
- Genus Iodamoeba Dobell 1919
- Genus Mastigamoeba Schulze 1875
- Family Rhizomastigidae Cavalier-Smith 2013
- Suborder Pelomyxina Starobogatov 1980
- Family Tricholimacidae Cavalier-Smith 2013
References
- Williams BP, Keeling PJ (2003-12-09). Littlewood T (ed.). The Evolution of Parasitism - A Phylogenetic Perspective. pp. 30–31. ISBN 9780080493749. Retrieved 20 February 2018.
- Ptáčková, Eliška; Kostygov, Alexei Yu; Chistyakova, Lyudmila V; Falteisek, Lukáš; Frolov, Alexander O; Patterson, David J; Walker, Giselle; Cepicka, Ivan (2013). "Evolution of Archamoebae: Morphological and Molecular Evidence for Pelobionts Including Rhizomastix, Entamoeba, Iodamoeba, and Endolimax". Protist. 164 (3): 380–410. doi:10.1016/j.protis.2012.11.005. PMID 23312407.
- Cavalier-Smith T (August 1998). "A revised six-kingdom system of life". Biol Rev Camb Philos Soc. 73 (3): 203–66. doi:10.1111/j.1469-185X.1998.tb00030.x. PMID 9809012.
- Keeling PJ (1998). "A kingdom's progress: Archezoa and the origin of eukaryotes" (PDF). BioEssays. 20: 87–95. doi:10.1002/(sici)1521-1878(199801)20:1<87::aid-bies12>3.0.co;2-4. Retrieved 20 February 2018.
- Cavalier-Smith T, Chao EE, Lewis R (June 2016). "187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution". Molecular Phylogenetics and Evolution. 99: 275–296. doi:10.1016/j.ympev.2016.03.023. PMID 27001604.
- Silar P (2016). "Protistes Eucaryotes: Origine, Evolution et Biologie des Microbes Eucaryotes" (PDF). HAL archives-ouvertes. pp. 1–462. ISBN 978-2-9555841-0-1.
- Kang S, Tice AK, Spiegel FW, Silberman JD, Pánek T, Cepicka I, et al. (September 2017). "Between a Pod and a Hard Test: The Deep Evolution of Amoebae". Molecular Biology and Evolution. 34 (9): 2258–2270. doi:10.1093/molbev/msx162. PMC 5850466. PMID 28505375.
- Pánek T, Zadrobílková E, Walker G, Brown MW, Gentekaki E, Hroudová M, et al. (May 2016). "First multigene analysis of Archamoebae (Amoebozoa: Conosa) robustly reveals its phylogeny and shows that Entamoebidae represents a deep lineage of the group". Molecular Phylogenetics and Evolution. 98: 41–51. doi:10.1016/j.ympev.2016.01.011. PMID 26826602.