Placozoa
The Placozoa are a basal form of free-living (non-parasitic) multicellular organism.[1] They are the simplest in structure of all animals. Three genera have been found: the classical Trichoplax adhaerens, Hoilungia hongkongensis, and Polyplacotoma mediterranea, where the last appears most basal. The last two have been found only since 2017.[2][3][4][5][6] Although the Placozoa were first discovered in 1883 by the German zoologist, Franz Eilhard Schulze (1840–1921)[7][8] and since the 1970s more systematically analyzed by the German protozoologist, Karl Gottlieb Grell (1912–1994),[9] a common name does not yet exist for the taxon; the scientific name means "flat animals".[10]
| Placozoa | |
|---|---|
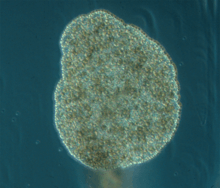 | |
| Trichoplax adhaerens | |
| Scientific classification | |
| Kingdom: | Animalia |
| Subkingdom: | Eumetazoa |
| Clade: | ParaHoxozoa |
| Phylum: | Placozoa Grell, 1971 |
| Type species | |
| Trichoplax adhaerens | |
| Families | |
Biology
Trichoplax is a small, flattened, animal around 1 mm (0.039 in) across. Like an Amoeba, it has no regular outline, although the lower surface is somewhat concave, and the upper surface is always flattened. The body consists of an outer layer of simple epithelium enclosing a loose sheet of stellate cells resembling the mesenchyme of some more complex animals. The epithelial cells bear cilia, which the animal uses to help it creep along the seafloor.[8]
The lower surface engulfs small particles of organic detritus, on which the animal feeds. It reproduces asexually, budding off smaller individuals, and the lower surface may also bud off eggs into the mesenchyme.[8]
Sexual reproduction has been reported to occur in one clade of the placozoa.[11][12] Intergenic recombination was observed as well as other hallmarks of sexual reproduction.
Some Trichoplax species contain Rickettsiales endosymbionts.[13] One of the at least 20 described species turned out to have two bacterial endosymbionts; Grellia which lives in the animal's endoplasmic reticulum and is assumed to play a role in the protein and membrane production. The other endosymbiont is the first described Margulisbacteria, that lives inside cells used for algal digestion. It appears to eat the fats and other lipids of the algae and provide its host with vitamins and amino acids in return.[14]
Evolutionary relationships
There is no convincing fossil record of the placozoa, although the Ediacaran biota (Precambrian, 550 million years ago) organism Dickinsonia may be allied with this phylum.[15]
Traditionally, classification was based on their level of organization: i.e. they possess no tissues or organs. However this may be as a result of secondary loss, so is inadequate to demark a clade. More recent work has attempted to classify them based on the DNA sequences in their genome; this has placed the phylum between the sponges and the eumetazoa.[16] In such a feature-poor phylum, molecular data are considered to provide the most reliable approximation of the placozoans' phylogeny.
Functional-morphology hypothesis
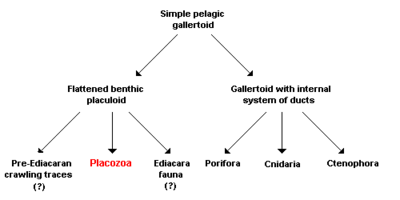
On the basis of their simple structure, the Placozoa were frequently viewed as a model organism for the transition from unicellular organisms to the multicellular animals (Metazoa) and are thus considered a sister taxon to all other metazoans:
| Metazoa |
| ||||||||||||
According to a functional-morphology model, all or most animals are descended from a gallertoid, a free-living (pelagic) sphere in seawater, consisting of a single ciliated layer of cells supported by a thin, noncellular separating layer, the basal lamina. The interior of the sphere is filled with contractile fibrous cells and a gelatinous extracellular matrix. Both the modern Placozoa and all other animals then descended from this multicellular beginning stage via two different processes:
- Infolding of the epithelium led to the formation of an internal system of ducts and thus to the development of a modified gallertoid from which the sponges (Porifera), Cnidaria and Ctenophora subsequently developed.
- Other gallertoids, according to this model, made the transition over time to a benthic mode of life; that is, their habitat has shifted from the open ocean to the floor (benthic zone). While the probability of encountering food, potential sexual partners, or predators is the same in all directions for animals floating freely in the water, there is a clear difference on the seafloor between the sides facing toward and away from the substrate, and between their orientation and the vertical direction perpendicular to the substrate. This results naturally in a selective advantage for flattening of the body, as of course can be seen in many benthic species. In the proposed functional-morphology model, the Placozoa, and possibly also several organisms known only from the fossil state, are descended from such a life form, which is now termed placuloid. Three different life strategies have accordingly led to three different lines of development:
- Animals that live interstitially in the sand of the ocean floor were responsible for the fossil crawling traces that are considered the earliest evidence of animals and are detectable even prior to the dawn of the Ediacaran Period in geology. These are usually attributed to bilaterally symmetrical worms, but the hypothesis presented here views animals derived from placuloids, and thus close relatives of Trichoplax adhaerens, to be the producers of the traces.
- Animals that incorporated algae as photosynthetically active endosymbionts, i.e. primarily obtaining their nutrients from their partners in symbiosis, were accordingly responsible for the mysterious creatures of the Ediacara fauna that are not assigned to any modern animal taxon and lived during the Ediacaran Period, before the start of the Paleozoic. Recent work has shown that some of the Ediacaran assemblages (e.g. Mistaken Point) were in deep water, below the photic zone, and that the organisms were not dependent on endosymbiotic photosynthesisers.
- Animals that grazed on algal mats were ultimately the direct ancestors of the Placozoa. The advantages of an amoeboid multiplicity of shapes thus allowed a previously present basal lamina and a gelatinous extracellular matrix to be lost secondarily. Pronounced differentiation between the ventral surface facing the substrate and the dorsal, facing away from it, accordingly led to the physiologically distinct cell layers of Trichoplax adhaerens that can still be seen today. Consequently, these are analogous, but not homologous, to ectoderm and endoderm, the "external" and "internal" cell layers in eumetazoans; i.e. the structures corresponding functionally to one another have, according to the proposed hypothesis, no common evolutionary origin.
Should the analysis presented above turn out to be correct, Trichoplax adhaerens would be the oldest branch of the multicellular animals and a relic of the Ediacara fauna, or even the pre-Ediacara fauna. Due to the absence of extracellular matrix and basal lamina, the development potential of these animals, very successful in their ecological niche, was of course limited, which would explain the low rate of evolution, referred to as bradytely, of their phenotype, their outward form as adults.
This hypothesis was supported by a recent analysis of the Trichoplax adhaerens mitochondrial genome in comparison to those of other animals,[17] The hypothesis was, however, rejected in a statistical analysis of the Trichoplax adhaerens whole genome sequence in comparison to the whole genome sequences of six other animals and two related non-animal species, but only at the p=0.07 level, which indicates a marginal level of statistical significance.[16]
Epitheliozoa hypothesis
A concept based on purely morphological characteristics pictures the Placozoa as the nearest relative of the animals with true tissues (Eumetazoa). The taxon they share, called the Epitheliozoa, is itself construed to be a sister group to the sponges (Porifera):
| Metazoa |
| ||||||||||||
The above view could be correct, although there is some evidence that the ctenophores, traditionally seen as Eumetazoa, may be the sister to all animals.[18] This is now a disputed classification.
The principal support for such a relationship comes from special cell/cell junctions, the belt desmosomes, that occur not just in the Placozoa but in all animals except the sponges; they enable the cells to join together in an unbroken layer like the epitheloid of the Placozoa. Trichoplax adhaerens also shares the ventral gland cells with most eumetazoans. Both characteristics can be considered apomorphies, i.e. evolutionarily derived features, and thus form the basis of a common taxon for all animals that possess them.
One possible scenario inspired by the proposed hypothesis starts with the idea that the monociliated cells of the epitheloid in Trichoplax adhaerens evolved by reduction of the collars in the collar cells (choanocytes) of sponges as the ancestors of the Placozoa abandoned a filtering mode of life. The epitheloid would then have served as the precursor to the true epithelial tissue of the eumetazoans.
In contrast to the model based on functional morphology described earlier, in the Epitheliozoa concept the ventral and dorsal cell layers of the Placozoa are homologs of endoderm and ectoderm, the two basic embryonic cell layers of the eumetazoans — the digestive gastrodermis in the Cnidaria or the gut epithelium in the bilaterally symmetrical Bilateria may have developed from endoderm, whereas ectoderm is, among other things, the precursor to the external skin layer (epidermis). The interior space pervaded by a fiber syncytium in the Placozoa would then correspond to connective tissue in the other animals. It is uncertain whether the calcium ions stored in the syncytium are related to the lime skeletons of many cnidarians.
As noted above, this hypothesis was supported in a statistical analysis of the Trichoplax adhaerens whole genome sequence in comparison to the whole genome sequences of six other animals and two related non-animal species.[16]
Eumetazoa hypothesis
A third hypothesis, based primarily on molecular genetics, views the Placozoa as highly simplified eumetazoans. According to this, Trichoplax adhaerens is descended from considerably more complex animals that already had muscles and nerve tissues. Both tissue types, as well as the basal lamina of the epithelium, were accordingly lost more recently by radical secondary simplification.
Various studies in this regard so far yield differing results for identifying the exact sister group: in one case the Placozoa would qualify as the nearest relatives of the Cnidaria, while in another they would be a sister group to the Ctenophora, and occasionally they are placed directly next to the Bilateria. Currently, they are typically placed according to the cladogram below:
| Metazoa |
| ||||||||||||||||||||||||
In this cladogram the Epitheliozoa and Eumetazoa are synonyms to each other and to the Diploblasts, and the Ctenophora are basal to them.
An argument raised against the proposed scenario is that it leaves morphological features of the animals completely out of consideration. The extreme degree of simplification that would have to be postulated for the Placozoa in this model, moreover, is known only for parasitic organisms but would be difficult to explain functionally in a free-living species like Trichoplax adhaerens.
This version is supported by statistical analysis of the Trichoplax adhaerens whole genome sequence in comparison to the whole genome sequences of six other animals and two related non-animal species. However, ctenophora was not included in the analyses, placing the placozoas outside of the sampled Eumetazoans.[16][19]
Cnidaria-sister hypothesis
DNA studies suggests that these organisms are related to Cnidaria, derived from planula larva (as seen in some Cnidaria).[20] The Bilateria also are derived from planuloids.[21][22][23][24][25][26][27][28] The Cnidaria and Placozoa body axis are overtly similar, and Placozoan and Cnidarian cells are responsive to the same neuropeptide antibodies despite extant Placozoa not developing any neurons.[29][30]
| Choanozoa |
| ||||||||||||||||||||||||||||||
| 950 mya |
References
- Placozoa at the US National Library of Medicine Medical Subject Headings (MeSH)
- Voigt, O; Collins AG; Pearse VB; Pearse JS; Hadrys H; Ender A (23 November 2004). "Placozoa — no longer a phylum of one". Current Biology. 14 (22): R944–5. doi:10.1016/j.cub.2004.10.036. PMID 15556848.
- Eitel, Michael; Osigus, Hans-Jürgen; DeSalle, Rob; Schierwater, Bernd (2 April 2013). "Global Diversity of the Placozoa". PLOS One. 8 (4): e57131. Bibcode:2013PLoSO...857131E. doi:10.1371/journal.pone.0057131. PMC 3614897. PMID 23565136.
- Eitel, Michael; Francis, Warren; Osigus, Hans-Jürgen; Krebs, Stefan; Vargas, Sergio; Blum, Helmut; Williams, Gray Argust; Schierwater, Bernd; Wörheide, Gert (2017-10-13). "A taxogenomics approach uncovers a new genus in the phylum Placozoa". bioRxiv: 202119. doi:10.1101/202119.
- Francis, Warren R.; Wörheide, Gert (2017-06-01). "Similar Ratios of Introns to Intergenic Sequence across Animal Genomes". Genome Biology and Evolution. 9 (6): 1582–1598. doi:10.1093/gbe/evx103. PMC 5534336. PMID 28633296.
- Schierwater, Bernd; Kamm, Kai; Herzog, Rebecca; Rolfes, Sarah; Osigus, Hans-Jürgen (2019-03-04). "Polyplacotoma mediterranea is a new ramified placozoan species". Current Biology. 29 (5): R148–R149. doi:10.1016/j.cub.2019.01.068. ISSN 0960-9822. PMID 30836080.
- F. E. Schulze "Trichoplax adhaerens n. g., n. s.", Zoologischer Anzeiger (Elsevier, Amsterdam and Jena) 6 (1883), p. 92.
- Barnes, Robert D. (1982). Invertebrate Zoology. Philadelphia: Holt-Saunders International. pp. 84–85. ISBN 978-0-03-056747-6.
- Grell, K. G. (1971). "Trichoplax adhaerens, F. E. Schulze und die Entstehung der Metazoen". Naturwissenschaftliche Rundschau. 24: 160.
- Rüdiger Wehner & Walter Gehring (June 2007). Zoologie (in German) (24th ed.). Stuttgart: Thieme. p. 696.
- Signorovitch AY, Dellaporta SL, Buss LW (2005). "Molecular signatures for sex in the Placozoa". Proceedings of the National Academy of Sciences of the United States of America. 102 (43): 15518–22. Bibcode:2005PNAS..10215518S. doi:10.1073/pnas.0504031102. PMC 1266089. PMID 16230622.
- Charlesworth D (2006). "Population genetics: using recombination to detect sexual reproduction: the contrasting cases of Placozoa and C. elegans". Heredity (Edinb). 96 (5): 341–2. doi:10.1038/sj.hdy.6800809. PMID 16552431.
- Kamm, Kai; Schierwater, Bernd; DeSalle, Rob (2019-01-05). "Innate immunity in the simplest animals – placozoans". BMC Genomics. 20 (1): 5. doi:10.1186/s12864-018-5377-3. ISSN 1471-2164. PMC 6321704. PMID 30611207.
- Deceptively simple: Minute marine animals live in a sophisticated symbiosis with bacteria - Phys.org
- Sperling, Erik; Vinther, Jakob; Pisani, Davide; Peterson, Kevin (2008). "A placozoan affinity for Dickinsonia and the evolution of Late Precambrian metazoan feeding modes" (PDF). In Cusack, M; Owen, A; Clark, N (eds.). Programme with Abstracts. Palaeontological Association Annual Meeting. 52. Glasgow, UK. p. 81.
- Srivastava, M.; Begovic, Emina; Chapman, Jarrod; Putnam, Nicholas H.; Hellsten, Uffe; Kawashima, Takeshi; Kuo, Alan; Mitros, Therese; et al. (21 August 2008). "The Trichoplax genome and the nature of placozoans" (PDF). Nature. 454 (7207): 955–960. Bibcode:2008Natur.454..955S. doi:10.1038/nature07191. PMID 18719581. Archived from the original (PDF) on 23 July 2018. Retrieved 2 September 2019.
- Dellaporta; Xu, A; Sagasser, S; Jakob, W; Moreno, MA; Buss, LW; Schierwater, B; et al. (6 June 2006). "'Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum'". Proceedings of the National Academy of Sciences of the United States of America. 103 (23): 8751–6. Bibcode:2006PNAS..103.8751D. doi:10.1073/pnas.0602076103. PMC 1470968. PMID 16731622.
- Whelan, Nathan V.; Kocot, Kevin M.; Moroz, Tatiana P.; Mukherjee, Krishanu; Williams, Peter; Paulay, Gustav; Moroz, Leonid L.; Halanych, Kenneth M. (2017-10-09). "Ctenophore relationships and their placement as the sister group to all other animals". Nature Ecology & Evolution. 1 (11): 1737–1746. doi:10.1038/s41559-017-0331-3. ISSN 2397-334X. PMC 5664179. PMID 28993654.
- Wallberg, Andreas; Thollesson, Mikael; Farris, James S.; Jondelius, Ulf (2004-12-01). "The phylogenetic position of the comb jellies (Ctenophora) and the importance of taxonomic sampling". Cladistics. 20 (6): 558–578. doi:10.1111/j.1096-0031.2004.00041.x. ISSN 1096-0031.
- Laumer, CE; Fernández, R; Lemer, S; Combosch, D; Kocot, KM; Riesgo, A; Andrade, SCS; Sterrer, W; Sørensen, MV; Giribet, G (1906). "Revisiting metazoan phylogeny with genomic sampling of all phyla". Proc Biol Sci. 286: 20190831.
- Aleshin VV, Petrov NB (2002) Molecular evidence of regression in evolution of metazoa. Zh Obshch Biol 63(3):195-208
- Laumer, Christopher E; Gruber-Vodicka, Harald; Hadfield, Michael G; Pearse, Vicki B; Riesgo, Ana; Marioni, John C; Giribet, Gonzalo (2018-10-30). "Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias". eLife. 7. doi:10.7554/elife.36278. ISSN 2050-084X. PMC 6277202. PMID 30373720.
- Syed, Tareq; Schierwater, Bernd (2002-06-01). "The evolution of the placozoa: A new morphological model". Senckenbergiana Lethaea. 82 (1): 315–324. doi:10.1007/bf03043791. ISSN 0037-2110.
- Hejnol, Andreas; Martindale, Mark Q. (2008-04-27). "Acoel development supports a simple planula-like urbilaterian". Philosophical Transactions of the Royal Society B: Biological Sciences. 363 (1496): 1493–1501. doi:10.1098/rstb.2007.2239. ISSN 0962-8436. PMC 2614228. PMID 18192185.
- Alzugaray, María Eugenia; Bruno, María Cecilia; Sambucaro, María José Villalobos; Ronderos, Jorge Rafael (2018-08-29). "The Evolutionary History of the Orexin/Allatotropin Gpcr Family: From Placozoa and Cnidaria to Vertebrata". bioRxiv: 403709. doi:10.1101/403709.
- Silva, Fernanda Britto da; Muschner, Valéria C.; Bonatto, Sandro L. (2007). "Phylogenetic position of Placozoa based on large subunit (LSU) and small subunit (SSU) rRNA genes". Genetics and Molecular Biology. 30 (1): 127–132. doi:10.1590/S1415-47572007000100022. ISSN 1415-4757.
- Adl, Sina M.; Bass, David; Lane, Christopher E.; Lukeš, Julius; Schoch, Conrad L.; Smirnov, Alexey; Agatha, Sabine; Berney, Cedric; Brown, Matthew W. (2018-09-26). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. ISSN 1066-5234. PMC 6492006. PMID 30257078.
- Giribet, Gonzalo; Edgecombe, Gregory D. (2020-03-03). The Invertebrate Tree of Life. Princeton University Press. ISBN 978-0-691-19706-7.
- DuBuc, Timothy Q; Ryan, Joseph; Martindale, Mark Q (2019-02-06). True, John (ed.). ""Dorsal-ventral" genes are part of an ancient axial patterning system: evidence from Trichoplax adhaerens (Placozoa)". Molecular Biology and Evolution. 36 (5): 966–973. doi:10.1093/molbev/msz025. ISSN 0737-4038. PMC 6501881. PMID 30726986.
- Schuchert, Peter (1993-03-01). "Trichoplax adhaerens (Phylum Placozoa) has Cells that React with Antibodies Against the Neuropeptide RFamide". Acta Zoologica. 74 (2): 115–117. doi:10.1111/j.1463-6395.1993.tb01227.x. ISSN 1463-6395.
External links
| Wikispecies has information related to Placozoa |
| Wikimedia Commons has media related to Placozoa. |
- The Trichoplax adhaerens Grell-BS-1999 v1.0 Genome Portal at the DOE Joint Genome Institute
- The Trichoplax Genome Project at the Yale Peabody Museum
- A Weird Wee Beastie: Trichoplax adhaerens
- Research articles from the ITZ, TiHo Hannover
- Information page from the University of California at Berkeley
- Ender A, Schierwater B (January 2003). "Placozoa are not derived cnidarians: evidence from molecular morphology". Mol. Biol. Evol. 20 (1): 130–4. doi:10.1093/molbev/msg018. PMID 12519915. – Mitochondrial DNA and 16S rRNA analysis and phylogeny of Trichoplax adhaerens
- Historical overview of Trichoplax research
- Science Daily:Genome Of Simplest Animal Reveals Ancient Lineage, Confounding Array Of Complex Capabilities
- Vicki Buchsbaum Pearse, and Oliver Voigt, 2007. "Field biology of placozoans (Trichoplax): distribution, diversity, biotic interactions. Integrative and Comparative Biology", doi:10.1093/icb/icm015.