Cyclin-dependent kinase 2
Cyclin-dependent kinase 2, also known as cell division protein kinase 2, or Cdk2, is an enzyme that in humans is encoded by the CDK2 gene.[5][6] The protein encoded by this gene is a member of the cyclin-dependent kinase family of Ser/Thr protein kinases. This protein kinase is highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, also known as Cdk1 in humans. It is a catalytic subunit of the cyclin-dependent kinase complex, whose activity is restricted to the G1-S phase of the cell cycle, where cells make proteins necessary for mitosis and replicate their DNA. This protein associates with and is regulated by the regulatory subunits of the complex including cyclin E or A. Cyclin E binds G1 phase Cdk2, which is required for the transition from G1 to S phase while binding with Cyclin A is required to progress through the S phase.[7] Its activity is also regulated by phosphorylation. Multiple alternatively spliced variants and multiple transcription initiation sites of this gene have been reported.[8] The role of this protein in G1-S transition has been recently questioned as cells lacking Cdk2 are reported to have no problem during this transition.[9]
Dispensability in normally functioning tissue
Original cell-culture based experiments demonstrated cell cycle arrest at the G1-S transition resulting from the deletion of Cdk2.[10] Later experiments showed that Cdk2 deletions lengthened the G1 phase of the cell cycle in mouse embryo fibroblasts. However, they still entered S phase after this period and were able to complete the remaining phases of the cell cycle.[11] When Cdk2 was deleted in mice, the animals remained viable despite a reduction in body size. However, meiotic function of both male and female mice was inhibited. This suggests that Cdk2 is non-essential for the cell cycle of healthy cells, but essential for meiosis and reproduction.[10] Cells in Cdk2 knockout mice likely undergo fewer divisions, contributing to the reduction in body size. Germ cells also stop dividing at prophase of meiosis, leading to reproductive sterility.[11] Cdk1 is now believed to compensate for many aspects of Cdk2 deletion, except for meiotic function.[10]
Mechanism of activation
Cyclin-dependent kinase 2 is structured in two lobes. The lobe beginning at the N-terminus (N-lobe) contains many beta sheets, while the C-terminus lobe (C-lobe) is rich in alpha helices.[7] Cdk2 is capable of binding to many different cyclins, including cyclins A, B, E, and possibly C.[10] Recent studies suggest Cdk2 binds preferentially to cyclins A and E, while Cdk1 prefers cyclins A and B.[12]

Cdk2 becomes active when a cyclin protein (either A or E) binds at the active site located between the N and C lobes of the kinase. Due to the location of the active site, partner cyclins interact with both lobes of Cdk2. Cdk2 contains an important alpha helix located in the C lobe of the kinase, called the C-helix or the PSTAIRE-helix. Hydrophobic interactions cause the C-helix to associate with another helix in the activating cyclin. Activation induces a conformational change where the helix rotates and moves closer to the N-lobe. This allows the glutamic acid located on the C-helix to form an ion pair with a nearby lysine side chain. The significance of this movement is that it brings the side chain of Glu 51 , which belongs to a triad of catalytic site residues conserved in all eukaryotic kinases, into the catalytic site. This triad (Lys 33, Glu 51 and Asp 145) is involved in ATP phosphate orientation and magnesium coordination, and is thought to be critical for catalysis. This conformational change also relocates the activation loop to the C-lobe, revealing the ATP binding site now available for new interactions. Finally, the Threonine-160 residue is exposed and phosphorylated as the C-lobe activation segment is displaced from the catalytic site and the threonine residue is no longer sterically hindered. The phosphorylated threonine residue creates stability in the final enzyme conformation. It is important to note that throughout this activation process, cyclins binding to Cdk2 do not undergo any conformational change.[14][7]

Role in DNA replication
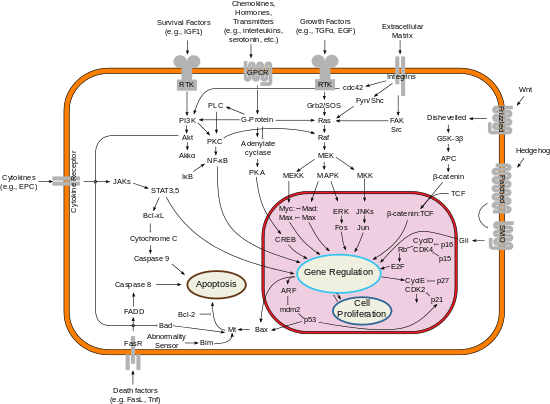
The success of the cell division process is dependent on the precise regulation of processes at both cellular and tissue levels. Complex interactions between proteins and DNA within the cell allow genomic DNA to be passed to daughter cells. Interactions between cells and extracellular matrix proteins allow new cells to be incorporated into existing tissues. At the cellular level, the process is controlled by different levels of cyclin-dependent kinases (Cdks) and their partner cyclins. Cells utilize various checkpoints as a means of delaying cell cycle progression until it can repair defects.[16]
Cdk2 is active during G1 and S phase of the cell cycle, and therefore acts as a G1-S phase checkpoint control. Prior to G1 phase, levels of Cdk4 and Cdk6 increase along with cyclin D. This allows for the partial phosphorylation of Rb, and partial activation of E2F at the beginning of G1 phase, which promotes cyclin E synthesis and increased Cdk2 activity. At the end of G1 phase, the Cdk2/Cyclin E complex reaches maximum activity and plays a significant role in the initiation of S phase.[17] Other non-Cdk proteins also become active during the G1-S phase transition. For example, the retinoblastoma (Rb) and p27 proteins are phosphorylated by Cdk2 – cyclin A/E complexes, fully deactivating them.[18] This allows E2F transcription factors to express genes that promote entry into S phase where DNA is replicated prior to division.[19][20][18] Additionally, NPAT, a known substrate of the Cdk2-Cyclin E complex, functions to activate histone gene transcription when phosphorylated.[21] This increases the synthesis of histone proteins (the major protein component of chromatin), and subsequently supports the DNA replication stage of the cell cycle. Finally, at the end of S phase, the ubiquitin proteasome degrades cyclin E.[11]
Cancer cell proliferation
Although Cdk2 is mostly dispensable in the cell cycle of normally functioning cells, it is critical to the abnormal growth processes of cancer cells. The CCNE1 gene produces cyclin E, one of the two major protein binding partners of Cdk2. Overexpression of CCNE1 occurs in many tumor cells, causing the cells to become dependent on Cdk2 and cyclin E.[12] Abnormal cyclin E activity is also observed in breast, lung, colorectal, gastric, and bone cancers, as well as in leukemia and lymphoma.[17] Likewise, abnormal expression of cyclin A2 is associated with chromosomal instability and tumor proliferation, while inhibition leads to decreased tumor growth.[22] Therefore, CDK2 and its cyclin binding partners represent possible therapeutic targets for new cancer therapeutics.[12] Pre-clinical models have shown preliminary success in limiting tumor growth, and have also been observed to reduce side effects of current chemotherapy drugs.[23][24][25]
Identifying selective Cdk2 inhibitors is difficult due to the extreme similarity between the active sites of Cdk2 and other Cdks, especially Cdk1.[12] Cdk1 is the only essential cyclin dependent kinase in the cell cycle, and inhibition could lead to unintended side effects.[26] Most CDK2 inhibitor candidates target the ATP binding site and can be divided into two main subclasses: type I and type II. Type I inhibitors competitively target the ATP binding site in its active state. Type II inhibitors target CDK2 in its unbound state, either occupying the ATP binding site or hydrophobic pocket within the kinase. Type II inhibitors are believed to be more selective.[24] Recently, the availability of new CDK crystal structures led to the identification of a potential allosteric binding site near the C-helix. Inhibitors of this allosteric site are classified as type III inhibitors.[27] Another possible target is the T-loop of CDK2. When cyclin A binds to CDK2, the N-terminal lobe rotates to activate the ATP binding site and switch the position of the activation loop, called the T-loop.[28]
Inhibitors
Known CDK inhibitors are p21Cip1 (CDKN1A) and p27Kip1 (CDKN1B).[29]
Drugs that inhibit Cdk2 and arrest the cell cycle, such as GW8510 and the experimental cancer drug seliciclib, may reduce the sensitivity of the epithelium to many cell cycle-active antitumor agents and, therefore, represent a strategy for prevention of chemotherapy-induced alopecia.[30]
Rosmarinic acid methyl ester is a plant-derived Cdk2 inhibitor, which was shown to suppress proliferation of vascular smooth muscle cells and to reduce neointima formation in mouse restenosis model.[31]
See also the PDB gallery below showing interactions with many inhibitors (inc Purvalanol B)
Gene regulation
In melanocytic cell types, expression of the CDK2 gene is regulated by the Microphthalmia-associated transcription factor.[32][33]
Interactions
Cyclin-dependent kinase 2 has been shown to interact with:
- BRCA1,[34][35][36]
- CDK2AP1,[37]
- CDKN1B[38][39][40][41][42]
- CDKN3,[43][44][45]
- CEBPA,[46]
- Cyclin A1,[47][48][49][50]
- Cyclin E1,[38][51][52][53][54][55]
- Flap structure-specific endonuclease 1,[56]
- ORC1L,[57]
- P21,[42][45][52][58][59]
- PPM1B,[60]
- PPP2CA,[60]
- Retinoblastoma-like protein 1,[51][61]
- Retinoblastoma-like protein 2,[51][62] and
- SKP2.[39][58][63]
References
- GRCh38: Ensembl release 89: ENSG00000123374 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025358 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Tsai LH, Harlow E, Meyerson M (September 1991). "Isolation of the human cdk2 gene that encodes the cyclin A- and adenovirus E1A-associated p33 kinase". Nature. 353 (6340): 174–7. doi:10.1038/353174a0. PMID 1653904.
- "Entrez Gene: CDK2 cyclin-dependent kinase 2".
- Echalier A, Endicott JA, Noble ME (March 2010). "Recent developments in cyclin-dependent kinase biochemical and structural studies". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1804 (3): 511–9. doi:10.1016/j.bbapap.2009.10.002. PMID 19822225.
- "Entrez Gene: CDK2 cyclin-dependent kinase 2".
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P (October 2003). "Cdk2 knockout mice are viable". Current Biology. 13 (20): 1775–85. doi:10.1016/j.cub.2003.09.024. PMID 14561402.
- Satyanarayana A, Kaldis P (August 2009). "Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms". Oncogene. 28 (33): 2925–39. doi:10.1038/onc.2009.170. PMID 19561645.
- Su TT, Stumpff J (March 2004). "Promiscuity rules? The dispensability of cyclin E and Cdk2". Science's STKE. 2004 (224): pe11. doi:10.1126/stke.2242004pe11. PMC 3242733. PMID 15026579.
- Wood DJ, Korolchuk S, Tatum NJ, Wang LZ, Endicott JA, Noble ME, Martin MP (November 2018). "Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition". Cell Chemical Biology. 26 (1): 121–130.e5. doi:10.1016/j.chembiol.2018.10.015. PMC 6344228. PMID 30472117.
- PDB: 1FIN; Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP (July 1995). "Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex". Nature. 376 (6538): 313–20. doi:10.1038/376313a0. PMID 7630397.
- Malumbres M (2014-06-30). "Cyclin-dependent kinases". Genome Biology. 15 (6): 122. doi:10.1186/gb4184. PMC 4097832. PMID 25180339.
- PDB: 1W98; Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, Johnson LN (February 2005). "The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles". The EMBO Journal. 24 (3): 452–63. doi:10.1038/sj.emboj.7600554. PMC 548659. PMID 15660127.
- Bartek J, Lukas C, Lukas J (October 2004). "Checking on DNA damage in S phase". Nature Reviews. Molecular Cell Biology. 5 (10): 792–804. doi:10.1038/nrm1493. PMID 15459660.
- Caruso JA, Duong MT, Carey JP, Hunt KK, Keyomarsi K (October 2018). "Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies". Cancer Research. 78 (19): 5481–5491. doi:10.1158/0008-5472.can-18-1235. PMC 6168358. PMID 30194068.
- Giacinti C, Giordano A (August 2006). "RB and cell cycle progression". Oncogene. 25 (38): 5220–7. doi:10.1038/sj.onc.1209615. PMID 16936740.
- Cobrinik D (April 2005). "Pocket proteins and cell cycle control". Oncogene. 24 (17): 2796–809. doi:10.1038/sj.onc.1208619. PMID 15838516.
- The molecular basis of cancer. Mendelsohn, John, 1936-, Gray, Joe W.,, Howley, Peter M.,, Israel, Mark A.,, Thompson, Craig (Craig B.) (Fourth ed.). Philadelphia, PA. 2015. ISBN 9781455740666. OCLC 870870610.CS1 maint: others (link)
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E (September 2000). "NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription". Genes & Development. 14 (18): 2283–97. doi:10.1101/gad.827700. PMC 316937. PMID 10995386.
- Gopinathan L, Tan SL, Padmakumar VC, Coppola V, Tessarollo L, Kaldis P (July 2014). "Loss of Cdk2 and cyclin A2 impairs cell proliferation and tumorigenesis". Cancer Research. 74 (14): 3870–9. doi:10.1158/0008-5472.CAN-13-3440. PMC 4102624. PMID 24802190.
- Xia P, Liu Y, Chen J, Coates S, Liu D, Cheng Z (October 2018). "Inhibition of cyclin-dependent kinase 2 protects against doxorubicin-induced cardiomyocyte apoptosis and cardiomyopathy". The Journal of Biological Chemistry. 293 (51): 19672–19685. doi:10.1074/jbc.ra118.004673. PMC 6314117. PMID 30361442.
- Whittaker SR, Mallinger A, Workman P, Clarke PA (May 2017). "Inhibitors of cyclin-dependent kinases as cancer therapeutics". Pharmacology & Therapeutics. 173: 83–105. doi:10.1016/j.pharmthera.2017.02.008. PMC 6141011. PMID 28174091.
- Cicenas J, Kalyan K, Sorokinas A, Stankunas E, Levy J, Meskinyte I, Stankevicius V, Kaupinis A, Valius M (June 2015). "Roscovitine in cancer and other diseases". Annals of Translational Medicine. 3 (10): 135. doi:10.3978/j.issn.2305-5839.2015.03.61. PMC 4486920. PMID 26207228.
- Brown NR, Korolchuk S, Martin MP, Stanley WA, Moukhametzianov R, Noble ME, Endicott JA (April 2015). "CDK1 structures reveal conserved and unique features of the essential cell cycle CDK". Nature Communications. 6: 6769. doi:10.1038/ncomms7769. PMC 4413027. PMID 25864384.
- Rastelli G, Anighoro A, Chripkova M, Carrassa L, Broggini M (2014-06-09). "Structure-based discovery of the first allosteric inhibitors of cyclin-dependent kinase 2". Cell Cycle. 13 (14): 2296–305. doi:10.4161/cc.29295. PMC 4111683. PMID 24911186.
- Pellerano M, Tcherniuk S, Perals C, Ngoc Van TN, Garcin E, Mahuteau-Betzer F, Teulade-Fichou MP, Morris MC (August 2017). "Targeting Conformational Activation of CDK2 Kinase". Biotechnology Journal. 12 (8): 1600531. doi:10.1002/biot.201600531. PMID 28430399.
- Levkau B, Koyama H, Raines EW, Clurman BE, Herren B, Orth K, Roberts JM, Ross R (March 1998). "Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade". Molecular Cell. 1 (4): 553–63. doi:10.1016/S1097-2765(00)80055-6. PMID 9660939.
- Davis ST, Benson BG, Bramson HN, Chapman DE, Dickerson SH, Dold KM, Eberwein DJ, Edelstein M, Frye SV, Gampe RT, Griffin RJ, Harris PA, Hassell AM, Holmes WD, Hunter RN, Knick VB, Lackey K, Lovejoy B, Luzzio MJ, Murray D, Parker P, Rocque WJ, Shewchuk L, Veal JM, Walker DH, Kuyper LF (January 2001). "Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors". Science. 291 (5501): 134–7. doi:10.1126/science.291.5501.134. PMID 11141566.
- Liu R, Heiss EH, Waltenberger B, Blažević T, Schachner D, Jiang B, Krystof V, Liu W, Schwaiger S, Peña-Rodríguez LM, Breuss JM, Stuppner H, Dirsch VM, Atanasov AG (April 2018). "Constituents of Mediterranean Spices Counteracting Vascular Smooth Muscle Cell Proliferation: Identification and Characterization of Rosmarinic Acid Methyl Ester as a Novel Inhibitor". Molecular Nutrition & Food Research. 62 (7): e1700860. doi:10.1002/mnfr.201700860. PMID 29405576.
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE (December 2004). "Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF". Cancer Cell. 6 (6): 565–76. doi:10.1016/j.ccr.2004.10.014. PMID 15607961.
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E (December 2008). "Novel MITF targets identified using a two-step DNA microarray strategy". Pigment Cell & Melanoma Research. 21 (6): 665–76. doi:10.1111/j.1755-148X.2008.00505.x. PMID 19067971.
- Chen Y, Farmer AA, Chen CF, Jones DC, Chen PL, Lee WH (July 1996). "BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner". Cancer Research. 56 (14): 3168–72. PMID 8764100.
- Ruffner H, Jiang W, Craig AG, Hunter T, Verma IM (July 1999). "BRCA1 is phosphorylated at serine 1497 in vivo at a cyclin-dependent kinase 2 phosphorylation site". Molecular and Cellular Biology. 19 (7): 4843–54. doi:10.1128/MCB.19.7.4843. PMC 84283. PMID 10373534.
- Wang H, Shao N, Ding QM, Cui J, Reddy ES, Rao VN (July 1997). "BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases". Oncogene. 15 (2): 143–57. doi:10.1038/sj.onc.1201252. PMID 9244350.
- Shintani S, Ohyama H, Zhang X, McBride J, Matsuo K, Tsuji T, Hu MG, Hu G, Kohno Y, Lerman M, Todd R, Wong DT (September 2000). "p12(DOC-1) is a novel cyclin-dependent kinase 2-associated protein". Molecular and Cellular Biology. 20 (17): 6300–7. doi:10.1128/MCB.20.17.6300-6307.2000. PMC 86104. PMID 10938106.
- Connor MK, Kotchetkov R, Cariou S, Resch A, Lupetti R, Beniston RG, Melchior F, Hengst L, Slingerland JM (January 2003). "CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis". Molecular Biology of the Cell. 14 (1): 201–13. doi:10.1091/mbc.E02-06-0319. PMC 140238. PMID 12529437.
- Rosner M, Hengstschläger M (November 2004). "Tuberin binds p27 and negatively regulates its interaction with the SCF component Skp2". The Journal of Biological Chemistry. 279 (47): 48707–15. doi:10.1074/jbc.M405528200. PMID 15355997.
- Youn CK, Cho HJ, Kim SH, Kim HB, Kim MH, Chang IY, Lee JS, Chung MH, Hahm KS, You HJ (February 2005). "Bcl-2 expression suppresses mismatch repair activity through inhibition of E2F transcriptional activity". Nature Cell Biology. 7 (2): 137–47. doi:10.1038/ncb1215. PMID 15619620.
- Porter LA, Kong-Beltran M, Donoghue DJ (September 2003). "Spy1 interacts with p27Kip1 to allow G1/S progression". Molecular Biology of the Cell. 14 (9): 3664–74. doi:10.1091/mbc.E02-12-0820. PMC 196558. PMID 12972555.
- Law BK, Chytil A, Dumont N, Hamilton EG, Waltner-Law ME, Aakre ME, Covington C, Moses HL (December 2002). "Rapamycin potentiates transforming growth factor beta-induced growth arrest in nontransformed, oncogene-transformed, and human cancer cells". Molecular and Cellular Biology. 22 (23): 8184–98. doi:10.1128/mcb.22.23.8184-8198.2002. PMC 134072. PMID 12417722.
- Yeh CT, Lu SC, Chao CH, Chao ML (May 2003). "Abolishment of the interaction between cyclin-dependent kinase 2 and Cdk-associated protein phosphatase by a truncated KAP mutant". Biochemical and Biophysical Research Communications. 305 (2): 311–4. doi:10.1016/s0006-291x(03)00757-5. PMID 12745075.
- Hannon GJ, Casso D, Beach D (March 1994). "KAP: a dual specificity phosphatase that interacts with cyclin-dependent kinases". Proceedings of the National Academy of Sciences of the United States of America. 91 (5): 1731–5. doi:10.1073/pnas.91.5.1731. PMC 43237. PMID 8127873.
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ (November 1993). "The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases". Cell. 75 (4): 805–16. doi:10.1016/0092-8674(93)90499-g. PMID 8242751.
- Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, Timchenko NA (October 2001). "C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4". Molecular Cell. 8 (4): 817–28. doi:10.1016/s1097-2765(01)00366-5. PMID 11684017.
- Sweeney C, Murphy M, Kubelka M, Ravnik SE, Hawkins CF, Wolgemuth DJ, Carrington M (January 1996). "A distinct cyclin A is expressed in germ cells in the mouse". Development. 122 (1): 53–64. PMID 8565853.
- Yang R, Morosetti R, Koeffler HP (March 1997). "Characterization of a second human cyclin A that is highly expressed in testis and in several leukemic cell lines". Cancer Research. 57 (5): 913–20. PMID 9041194.
- Müller-Tidow C, Wang W, Idos GE, Diederichs S, Yang R, Readhead C, Berdel WE, Serve H, Saville M, Watson R, Koeffler HP (April 2001). "Cyclin A1 directly interacts with B-myb and cyclin A1/cdk2 phosphorylate B-myb at functionally important serine and threonine residues: tissue-specific regulation of B-myb function". Blood. 97 (7): 2091–7. doi:10.1182/blood.v97.7.2091. PMID 11264176.
- Brown NR, Noble ME, Endicott JA, Johnson LN (November 1999). "The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases". Nature Cell Biology. 1 (7): 438–43. doi:10.1038/15674. PMID 10559988.
- Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E (February 1999). "Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest". Molecular and Cellular Biology. 19 (2): 1460–9. doi:10.1128/mcb.19.2.1460. PMC 116074. PMID 9891079.
- McKenzie PP, Danks MK, Kriwacki RW, Harris LC (July 2003). "P21Waf1/Cip1 dysfunction in neuroblastoma: a novel mechanism of attenuating G0-G1 cell cycle arrest". Cancer Research. 63 (13): 3840–4. PMID 12839982.
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM (September 1992). "Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle". Science. 257 (5077): 1689–94. doi:10.1126/science.1388288. PMID 1388288.
- Mayer C, Zhao J, Yuan X, Grummt I (February 2004). "mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability". Genes & Development. 18 (4): 423–34. doi:10.1101/gad.285504. PMC 359396. PMID 15004009.
- Boudrez A, Beullens M, Groenen P, Van Eynde A, Vulsteke V, Jagiello I, Murray M, Krainer AR, Stalmans W, Bollen M (August 2000). "NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry". The Journal of Biological Chemistry. 275 (33): 25411–7. doi:10.1074/jbc.M001676200. PMID 10827081.
- Henneke G, Koundrioukoff S, Hübscher U (July 2003). "Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation". Oncogene. 22 (28): 4301–13. doi:10.1038/sj.onc.1206606. PMID 12853968.
- Méndez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B (March 2002). "Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication". Molecular Cell. 9 (3): 481–91. doi:10.1016/s1097-2765(02)00467-7. PMID 11931757.
- Yam CH, Ng RW, Siu WY, Lau AW, Poon RY (January 1999). "Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2". Molecular and Cellular Biology. 19 (1): 635–45. doi:10.1128/mcb.19.1.635. PMC 83921. PMID 9858587.
- Ono T, Kitaura H, Ugai H, Murata T, Yokoyama KK, Iguchi-Ariga SM, Ariga H (October 2000). "TOK-1, a novel p21Cip1-binding protein that cooperatively enhances p21-dependent inhibitory activity toward CDK2 kinase". The Journal of Biological Chemistry. 275 (40): 31145–54. doi:10.1074/jbc.M003031200. PMID 10878006.
- Cheng A, Kaldis P, Solomon MJ (November 2000). "Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2C alpha and beta 2 isoforms". The Journal of Biological Chemistry. 275 (44): 34744–9. doi:10.1074/jbc.M006210200. PMID 10934208.
- Leng X, Noble M, Adams PD, Qin J, Harper JW (April 2002). "Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4". Molecular and Cellular Biology. 22 (7): 2242–54. doi:10.1128/mcb.22.7.2242-2254.2002. PMC 133692. PMID 11884610.
- Lacy S, Whyte P (May 1997). "Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes". Oncogene. 14 (20): 2395–406. doi:10.1038/sj.onc.1201085. PMID 9188854.
- Marti A, Wirbelauer C, Scheffner M, Krek W (May 1999). "Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation". Nature Cell Biology. 1 (1): 14–9. doi:10.1038/8984. PMID 10559858.
Further reading
- Kaldis P, Aleem E (November 2005). "Cell cycle sibling rivalry: Cdc2 vs. Cdk2". Cell Cycle. 4 (11): 1491–4. doi:10.4161/cc.4.11.2124. PMID 16258277.
- Moore NL, Narayanan R, Weigel NL (February 2007). "Cyclin dependent kinase 2 and the regulation of human progesterone receptor activity". Steroids. 72 (2): 202–9. doi:10.1016/j.steroids.2006.11.025. PMC 1950255. PMID 17207508.
External links
- Cyclin-Dependent+Kinase+2 at the US National Library of Medicine Medical Subject Headings (MeSH)
- CDK2 human gene location in the UCSC Genome Browser.
- CDK2 human gene details in the UCSC Genome Browser.