SH2 domain
The SH2 (Src Homology 2) domain is a structurally conserved protein domain contained within the Src oncoprotein[2] and in many other intracellular signal-transducing proteins.[3] SH2 domains allow proteins containing those domains to dock to phosphorylated tyrosine residues on other proteins. SH2 domains are commonly found in adaptor proteins that aid in the signal transduction of receptor tyrosine kinase pathways.[4]
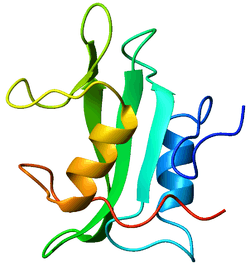 Crystallographic structure of the SH2 domain. The structure consists of a large beta sheet (green) flanked by two alpha-helices (orange and blue).[1] | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | SH2 | ||||||||
| Pfam | PF00017 | ||||||||
| InterPro | IPR000980 | ||||||||
| SMART | SH2 | ||||||||
| PROSITE | PDOC50001 | ||||||||
| SCOPe | 1sha / SUPFAM | ||||||||
| CDD | cd00173 | ||||||||
| |||||||||
Background
SH2 is conserved by signalization of protein tyrosine kinase, which are binding on phosphotyrosine (pTyr).[5] In the human proteome the class of pTyr-selective recognition domains is represented by SH2 domains. The N-terminal SH2 domains of cytoplasmic tyrosine kinase was at the beginning of evolution evolved with the occurrence of tyrosine phosphorylation. At the beginning it was supposed that, these domains serve as a substrate for their target kinase.[6]
Protein-protein interactions play a major role in cellular growth and development. Modular domains, which are the subunits of a protein, moderate these protein interactions by identifying short peptide sequences. These peptide sequences determine the binding partners of each protein. One of the more prominent domains is the SH2 domain. SH2 domains play a vital role in cellular communication. Its length is approximately 100 amino acids long and it is found within 111 human proteins.[7] Regarding its structure, it contains 2 alpha helices and 7 beta strands. Research has shown that it has a high affinity to phosphorylated tyrosine residues and it is known to identify a sequence of 3-6 amino acids within a peptide motif.
Binding and phosphorylation
SH2 domains typically bind a phosphorylated tyrosine residue in the context of a longer peptide motif within a target protein, and SH2 domains represent the largest class of known pTyr-recognition domains.[8][9]
Phosphorylation of tyrosine residues in a protein occurs during signal transduction and is carried out by tyrosine kinases. In this way, phosphorylation of a substrate by tyrosine kinases acts as a switch to trigger binding to an SH2 domain-containing protein. Many tyrosine containing short linear motifs that bind to SH2 domains are conserved across a wide variety of higher Eukaryotes.[10] The intimate relationship between tyrosine kinases and SH2 domains is supported by their coordinate emergence during eukaryotic evolution.
Diversity
SH2 domains are not present in yeast and appear at the boundary between protozoa and animalia in organisms such as the social amoeba Dictyostelium discoideum.[11]
A detailed bioinformatic examination of SH2 domains of human and mouse reveals 120 SH2 domains contained within 115 proteins encoded by the human genome,[12] representing a rapid rate of evolutionary expansion among the SH2 domains.
A large number of SH2 domain structures have been solved and many SH2 proteins have been knocked out in mice.
Function
The function of SH2 domains is to specifically recognize the phosphorylated state of tyrosine residues, thereby allowing SH2 domain-containing proteins to localize to tyrosine-phosphorylated sites. This process constitutes the fundamental event of signal transduction through a membrane, in which a signal in the extracellular compartment is "sensed" by a receptor and is converted in the intracellular compartment to a different chemical form, i.e. that of a phosphorylated tyrosine. Tyrosine phosphorylation leads to activation of a cascade of protein-protein interactions whereby SH2 domain-containing proteins are recruited to tyrosine-phosphorylated sites. This process initiates a series of events which eventually result in altered patterns of gene expression or other cellular responses. The SH2 domain, which was first identified in the oncoproteins Src and Fps, is about 100 amino-acid residues long. It functions as a regulatory module of intracellular signaling cascades by interacting with high affinity to phosphotyrosine-containing target peptides in a sequence-specific and strictly phosphorylation-dependent manner.
Applications
SH2 domains, and other binding domains, have been used in protein engineering to create protein assemblies. Protein assemblies are formed when several proteins bind to one another to create a larger structure (called a supramolecular assembly). Using molecular biology techniques, fusion proteins of specific enzymes and SH2 domains have been created, which can bind to each other to form protein assemblies.
Since SH2 domains require phosphorylation in order for binding to occur, the use of kinase and phosphatase enzymes gives researchers control over whether protein assemblies will form or not. High affinity engineered SH2 domains have been developed and utilized for protein assembly applications.[13]
The goal of most protein assembly formation is to increase the efficiency of metabolic pathways via enzymatic co-localization.[14] Other applications of SH2 domain mediated protein assemblies have been in the formation of high density fractal-like structures, which have extensive molecular trapping properties.[15]
Examples
Human proteins containing this domain include:
- ABL1; ABL2
- BCAR3; BLK; BLNK; BMX; BTK
- CHN2; CISH; CRK; CRKL; CSK
- DAPP1
- FER; FES; FGR; FRK; FYN
- GRAP; GRAP2; GRB10; GRB14; GRB2; GRB7
- HCK; HSH2D
- INPP5D; INPPL1; ITK; JAK2; LCK; LCP2; LYN
- MATK; NCK1; NCK2
- PIK3R1; PIK3R2; PIK3R3; PLCG1; PLCG2; PTK6; PTPN11; PTPN6; RASA1
- SH2B1; SH2B2; SH2B3; SH2D1A; SH2D1B; SH2D2A; SH2D3A; SH2D3C; SH2D4A; SH2D4B; SH2D5; SH2D6; SH3BP2; SHB; SHC1; SHC3; SHC4; SHD; SHE
- SLA; SLA2
- SOCS1; SOCS2; SOCS3; SOCS4; SOCS5; SOCS6; SOCS7
- SRC; SRMS
- STAT1; STAT2; STAT3; STAT4; STAT5A; STAT5B; STAT6
- SUPT6H; SYK
- TEC; TENC1; TNS; TNS1; TNS3; TNS4; TXK
- VAV1; VAV2; VAV3
- YES1; ZAP70
See also
- Phosphotyrosine-binding domains also bind phosphorylated tyrosines
- Anthony Pawson, discoverer of the SH2 Domain
- SH2 Domain website created by lab of Dr. Nash
References
- PDB: 1lkk; Tong L, Warren TC, King J, Betageri R, Rose J, Jakes S (March 1996). "Crystal structures of the human p56lck SH2 domain in complex with two short phosphotyrosyl peptides at 1.0 A and 1.8 A resolution". Journal of Molecular Biology. 256 (3): 601–10. doi:10.1006/jmbi.1996.0112. PMID 8604142.
- Sadowski I, Stone JC, Pawson T (December 1986). "A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps". Molecular and Cellular Biology. 6 (12): 4396–408. doi:10.1128/mcb.6.12.4396. PMC 367222. PMID 3025655.
- Russell RB, Breed J, Barton GJ (June 1992). "Conservation analysis and structure prediction of the SH2 family of phosphotyrosine binding domains". FEBS Letters. 304 (1): 15–20. doi:10.1016/0014-5793(92)80579-6. PMID 1377638.
- Koytiger G, Kaushansky A, Gordus A, Rush J, Sorger PK, MacBeath G (May 2013). "Phosphotyrosine signaling proteins that drive oncogenesis tend to be highly interconnected". Molecular & Cellular Proteomics. 12 (5): 1204–13. doi:10.1074/mcp.M112.025858. PMC 3650332. PMID 23358503.
- Chervitz SA, Aravind L, Sherlock G, Ball CA, Koonin EV, Dwight SS, Harris MA, Dolinski K, Mohr S, Smith T, Weng S, Cherry JM, Botstein D (December 1998). "Comparison of the complete protein sets of worm and yeast: orthology and divergence". Science. 282 (5396): 2022–8. doi:10.1126/science.282.5396.2022. PMC 3057080. PMID 9851918.
- Pawson T, Gish GD, Nash P (December 2001). "SH2 domains, interaction modules and cellular wiring". Trends in Cell Biology. 11 (12): 504–11. doi:10.1016/s0962-8924(01)02154-7. PMID 11719057.
- Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD (December 2011). "The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes". Science Signaling. 4 (202): ra83. doi:10.1126/scisignal.2002105. PMC 4255630. PMID 22155787.
- Pawson T, Gish GD, Nash P (December 2001). "SH2 domains, interaction modules and cellular wiring". Trends in Cell Biology. 11 (12): 504–11. doi:10.1016/S0962-8924(01)02154-7. PMID 11719057.
- Huang H, Li L, Wu C, Schibli D, Colwill K, Ma S, Li C, Roy P, Ho K, Songyang Z, Pawson T, Gao Y, Li SS (April 2008). "Defining the specificity space of the human SRC homology 2 domain". Molecular & Cellular Proteomics. 7 (4): 768–84. doi:10.1074/mcp.M700312-MCP200. PMID 17956856.
- Ren S, Yang G, He Y, Wang Y, Li Y, Chen Z (October 2008). "The conservation pattern of short linear motifs is highly correlated with the function of interacting protein domains". BMC Genomics. 9: 452. doi:10.1186/1471-2164-9-452. PMC 2576256. PMID 18828911.
- Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, et al. (May 2005). "The genome of the social amoeba Dictyostelium discoideum". Nature. 435 (7038): 43–57. doi:10.1038/nature03481. PMC 1352341. PMID 15875012.
- Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD (June 2006). "The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling". Molecular Cell. 22 (6): 851–68. doi:10.1016/j.molcel.2006.06.001. PMID 16793553.
- Kaneko, T.; Huang, H.; Cao, X.; Li, X.; Li, C.; Voss, C.; Sidhu, S. S.; Li, S. S. C. (2012-09-25). "Superbinder SH2 Domains Act as Antagonists of Cell Signaling". Science Signaling. 5 (243): ra68. doi:10.1126/scisignal.2003021. ISSN 1945-0877. PMID 23012655.
- Yang, Lu; Dolan, E.M.; Tan, S.K.; Lin, T.; Sontag, E.D.; Khare, S.D. (2017). "Computation-Guided Design of a Stimulus-Responsive Multienzyme Supramolecular Assembly". ChemBioChem. 18 (20): 2000–2006. doi:10.1002/cbic.201700425. ISSN 1439-7633. PMID 28799209.
- Hernández N.E., Hansen W.A., Zhu D., Shea M.E., Khalid M., Manichev V., Putnins M., Chen M., Dodge A.G., Yang L., Marrero-Berríos I., Banal M., Rechani P., Gustafsson T., Feldman L.C., Lee S-.H., Wackett L.P., Dai W., Khare S.D. (2019). Stimulus-responsive self-assembly of protein-based fractals by computational design. Nat. Chem. 2019 11(7): 605-614. Pre-print available at bioRxiv doi: 10.1101/274183.