Phosphotyrosine-binding domain
In molecular biology, Phosphotyrosine-binding domains are protein domains which bind to phosphotyrosine.
| Phosphotyrosine-binding domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
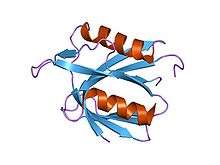 Structure of the PTB domain of tensin1.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | PTB | ||||||||||
| Pfam | PF08416 | ||||||||||
| InterPro | IPR013625 | ||||||||||
| CDD | cd00934 | ||||||||||
| |||||||||||
| PTB domain (IRS-1 type) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
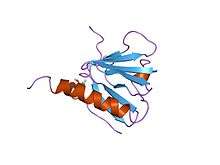 irs-1 ptb domain complexed with a il-4 receptor phosphopeptide, nmr, minimized average structure | |||||||||
| Identifiers | |||||||||
| Symbol | IRS | ||||||||
| Pfam | PF02174 | ||||||||
| InterPro | IPR002404 | ||||||||
| SMART | PTBI | ||||||||
| SCOPe | 1cli / SUPFAM | ||||||||
| CDD | cd01204 | ||||||||
| |||||||||
The phosphotyrosine-binding domain (PTB, also phosphotyrosine-interaction or PI domain) in the protein tensin tends to be found at the C-terminus. Tensin is a multi-domain protein that binds to actin filaments and functions as a focal-adhesion molecule (focal adhesions are regions of plasma membrane through which cells attach to the extracellular matrix). Human tensin has actin-binding sites, an SH2 (Pfam PF00017) domain and a region similar to the tumour suppressor PTEN.[2] The PTB domain interacts with the cytoplasmic tails of beta integrin by binding to an NPXY motif.[3]
The phosphotyrosine-binding domain of insulin receptor substrate-1 is not related to the phosphotyrosine-binding domain of tensin. Insulin receptor substrate-1 proteins contain both a pleckstrin homology domain and a phosphotyrosine binding (PTB) domain. The PTB domains facilitate interaction with the activated tyrosine-phosphorylated insulin receptor. The PTB domain is situated towards the N terminus. Two arginines in this domain are responsible for hydrogen bonding phosphotyrosine residues on an Ac-LYASSNPApY-NH2 peptide in the juxtamembrane region of the insulin receptor. Further interactions via "bridged" water molecules are coordinated by residues an Asn and a Ser residue.[4] The PTB domain has a compact, 7-stranded beta-sandwich structure, capped by a C-terminal helix. The substrate peptide fits into an L-shaped surface cleft formed from the C-terminal helix and strands 5 and 6.[5]
Human proteins containing these domains
APBA1; APBA2; APBA3; APPL1; EPS8; EPS8L1; EPS8L2; EPS8L3; TENC1; TNS; TNS1; TNS3; TNS4; DOK1; DOK2; DOK3; DOK4; DOK5; DOK6; DOK7; FRS2; FRS3; IRS1; IRS2; IRS4; NOS1AP; TLN1; TLN2
See also
- SH2 domains also bind phosphorylated tyrosines
References
- McCleverty CJ, Lin DC, Liddington RC (June 2007). "Structure of the PTB domain of tensin1 and a model for its recruitment to fibrillar adhesions". Protein Sci. 16 (6): 1223–9. doi:10.1110/ps.072798707. PMC 2206669. PMID 17473008.
- Chen H, Ishii A, Wong WK, Chen LB, Lo SH (October 2000). "Molecular characterization of human tensin". Biochem. J. 351 (2): 403–11. doi:10.1042/0264-6021:3510403. PMC 1221376. PMID 11023826.
- Lo SH (January 2004). "Tensin". Int. J. Biochem. Cell Biol. 36 (1): 31–4. doi:10.1016/S1357-2725(03)00171-7. PMID 14592531.
- Eck MJ, Dhe-Paganon S, Trub T, Nolte RT, Shoelson SE (May 1996). "Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor". Cell. 85 (5): 695–705. doi:10.1016/S0092-8674(00)81236-2. PMID 8646778.
- Zhou MM, Huang B, Olejniczak ET, Meadows RP, Shuker SB, Miyazaki M, Trub T, Shoelson SE, Fesik SW (April 1996). "Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain". Nat. Struct. Biol. 3 (4): 388–93. doi:10.1038/nsb0496-388. PMID 8599766.