L1 (protein)
L1, also known as L1CAM, is a transmembrane protein member of the L1 protein family, encoded by the L1CAM gene. This protein, of 200-220 kDa, is a neuronal cell adhesion molecule with a strong implication in cell migration, adhesion, neurite outgrowth, myelination and neuronal differentiation.[5] It also plays a key role in treatment-resistant cancers due to its function. It was first identified in 1984 by M. Schachner who found the protein in post-mitotic mice neurons.
Mutations in the L1 protein are the cause of L1 syndrome, sometimes known by the acronym CRASH (corpus callosum hypoplasia, retardation, aphasia, spastic paraplegia and hydrocephalus).[6]
Tissue and cellular distribution
L1 protein is located all over the nervous system on the surface of neurons. It is placed along the cellular membrane so that one end of the protein remains inside the nerve cell while the other end stays on the outer surface of the neurone. This position allows the protein to activate chemical signals which spread through the neurone.[7]
There are a wide variety of cells which express the protein L1, not only neuronal cells but also some non-neuronal ones. Cells which are known nowadays to express the protein L1 are: immature oligodendrocytes and Schwann cells, which are non-neuronal cells that provide support and protection for neurons and form myelin; T cells which are lymphocytes involved in cell-mediated immunity; other types of lymphocytes such as B cells and Monocytes. It is also expressed in intestinal epithelial progenitor cells, cerebellum neurons such as Cerebellum granule cell and Purkinje cells. Finally, it is expressed in multiple tumor cells for example Melanoma and lung carcinoma cells.[5]
Gene
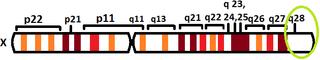
The human L1CAM gene is found in X chromosome regions that are implicated in different neuromuscular diseases, and near the one associated with mental retardation. L1CAM gene is located in the long arm of X chromosome in Xq28 position.[8][9]
Structure
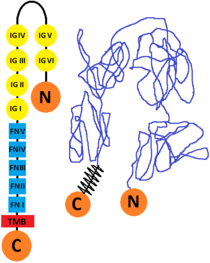
The L1 cell adhesion molecule (L1CAM) is a cell surface glycoprotein found in humans (and other forms of life as mice, for example) which has a 1253 amino acid protein sequence. The extracellular portion is formed of six immunoglobulin domains followed by five fibronectin type III domains which are connected to a small intracellular domain by a transmembrane helix. The human protein is very similar to the one that is found in mice (they are 92% identical at amino acid level, this enabling the scientists to study its structure. There are other CAM proteins like Ng-CAM (found in chicken) which has lower similarities to the human one (they are 40% identical at the amino acid level). The comparative of the sequences from human, mouse, chick and Drosophila and its good conservation, indicates that the L1 immunoglobulin domain 2 and fibronectin type III domain 2 probably are functionally important.[10][11]
Function
L1 is an important protein for the development of the nervous system affecting both cell adhesion and motility.
Cell adhesion
L1 has a static function as a cell adhesion molecule which connects different cells. It is involved in the adhesion between neurons and in the growth and association of neurites called neurite fasciculation.[12]
Cell motility
Motility promoting functions are related to the regulation of the movement of nerve cells during neural development. L1 is present in developing neurons and plays an important role in guiding new neurons into the correct positions and helping axons grow and make connections with other neurons. L1 is also involved in synaptic plasticity, which is the ability of synapses to strengthen or weaken, and it also plays a role in regeneration after trauma.
Some studies have proved that L1 has a role in tumor growth, tumor cell invasion, metastasis of melanoma, ovarian and colon cancer[13] due to an overexpression of the protein L1 that improves cell motion of the malignant cells.
The domains of this protein promote homophilic interactions, where adhesion molecules on one cell interact with identical molecules on the other cell. And also heterophilic interactions, where an adhesion molecule on one cell works as a receptor that connects with a different molecule on the other cell.[14][15] These interactions promote cell adhesion and regulation of signal transduction.
In addition, L1 participates in myelination processes, which are involved in the proliferation of myelin through the nervous system (specifically the progressive myelination of nerve axon fibers), by mediating the elongation of Schwann cells along the axon.
Nervous system
L1 is involved in neuron-neuron adhesion, neurite fasciculation, outgrowth of neurites, cerebellar granule cell migration, neurite outgrowth on Schwann cells and interactions among epithelial cells of intestinal crypts.[16] As a consequence, mutations in the L1CAM gene cause the Nervous System to malfunction. The main disorders linked to this mutation are known by the acronym CRASH or can be also referred as L1 syndrome. This includes disorders such as HSAS, MASA syndrome, agenesis of the corpus callosum and spastic paraplegia. Lower limb spasticity, mental retardation, hydrocephalus and flexion deformity of the thumbs are some of the symptoms expressed mostly in male individuals who suffer from this condition.[17][18][19] Although the pathological mechanisms leading to L1 syndrome are still unknown, about 200 mutations of the L1CAM gene have been identified and then associated with the syndrom. These mutations mostly affect structurally important key residues in the extracellular region of L1 causing alterations in the protein binding properties, which correlate to the impairment of neuronal physiological mechanisms such as cell adhesion or specific interacting with other molecules.[20] Ankyrin interaction with L1CAM is an example of a protein binding that fails in CRASH patients[21] due to a mutation that causes leucine and histidine to replace serine and tyrosine respectively, in the SFIGQY motif, where ankyrin should be bound in the L1CAM family cytoplasmic terminus.[22][23] Ankyrin-L1CAM interaction is involved in the growth cone initiation, consequently, a failure in this interaction causes neurites to not reach synaptic target.
Furthermore, evidence shows there is a correlation between fetal alcohol spectrum disorder and L1 protein since ethanol inhibits L1-mediated adhesion and neurite outgrowth.[24] Hirschsprung's disease has also been linked to a L1CAM malfunction.[25]
Transcription and synthesis
The gene that regulates L1CAM transcription is found in chromosome X. The L1CAM gene is 24,657 bp in length, and is made up of 28 exons. The alternative splicing of this gene leads to multiple transcript variants (there are 7 different transcripts of the gene),[26] including some that have an alternate exon that is considered to be specific to neurons.[27] L1 transcription is known to take place in human fetal brain and in neuroblastoma and retinoblastoma cell lines. L1 is also expressed in the rhabdomyosarcoma cell lines RD and A-204. Two forms of L1 can be found in humans, with the difference that one has a 12-bp cytoplasmic segment and the other lacks of it.[28] The regulation of L1CAM expression in transcription is not fully comprehended. Two sites were verified in endometrial carcinoma cell lines and seem to be used in a specific manner depending of the cell type. There are two transcription beginning sites, located in two different exons (in front of a non-translated exon 0 and next to the first protein-coding exon 1).[29] SLUG (SNAI2), a transcription factor, upregulates the expression of L1CAM.[30]
Sequences and different isoforms
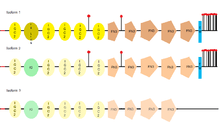
L1CAM has three different isoforms, that differ in their amino acid sequency, because of alternative splicing (a process that allows obtaining different mRNA mature molecules from one primary transcript of mRNA). L1CAM isoform 1 is known as the canonical sequence.[31] The main difference between them is where they can be found, for example, the full-length isoform (isoform 1), is the one usually found in neural cells, while the short one or nonneural isoform (isoform 2), is predominant in the other cell types.[32]
| Length (n aa) | Mass (Da) | Sequence | |
|---|---|---|---|
| Isoform 1
(fl-L1) |
1,257 | 140,003 | Canonical sequence. |
| Isoform 2
(sh-L1) |
1,253 | 139,517 | Differs from the canonical sequence in the amino acids between position 1177 and 1180, which aren't found in this isoform. |
| Isoform 3 | 1,248 | 138,908 | Differs from the canonical sequence in the amino acids between position 26 and 31 where six amino acids are exchanged for a leucine and as the previous one, in the amino acids between position 1177 and 1180, which aren't found in this isoform.[33] |
Interactions
L1 (protein) has been shown to interact with NUMB.[34]
Ig-like domain interactions
L1CAM is capable of folding into a horseshoe configuration by the establishment of homophilic interactions within Ig-like domains of the same protein (the first and the second Ig motifs folding back onto the 4th and 3rd motifs). This conformation is essential for L1CAM being able to interact with other molecules and subsequently performing some of its most important functions.
Ig-like domains are implicated in many homophilic interactions with other L1CAM proteins located in adjacent cells. L1CAM molecules interact via the Ig (1-4)-like domains, allowing cell to cell adhesion. They are also important in the formation of heterophilic interactions with NCAM, TAG-1, F11 and receptor tyrosine kinases (specially during the development of the nervous system).
The six Ig motif of the L1 protein contains an Arg-Gly-Asp sequence which allows binding with diverse surface cell integrins. This interaction leads to a signaling cascade which activates focal adhesion kinases (FAK) which are then converted to its active state and form the FAK/SRC complex. The latest functions as an activator of mitogen-activated protein kinases. Another function derived from integrin binding is the activation of NF-κB which results in making cells more motile and invasive.[5]
Fibronectin domain interactions
Fibronectin domains of L1 protein are also capable of binding cell surface integrins. They interact with fibroblast growth factor receptor 1, which suggests it may be linked to the modulating of neuronal differentiation.[5]
Cytoplasmic tail interactions
The most important binding partners of the cytoplasmic tail of L1 proteins are ankyrins. The interaction is held in high-affinity binding sites located within the so-called “ank repeats” also known as membrane-binding domains.[5] This interaction allows L1 protein connect with the cell's cytoskeleton. Also, L1 protein cytoplasmic tail can bind adaptor 2 (ADP), a key component of clathrin mediated endocytosis.
The fact this region contains some phosphorylation sites suggests L1 may be subject to regulation by kinases.[5]
Implications in cancer metastasis
L1CAM protein expression is normally restricted to neurons. However, it has been noticed there's L1CAM overexpression in all types of cancer cells, which has been associated with poor prognosis, tumor progression and metastasis.[35] This up-regulation may not be necessarily associated with mutations in L1 transcription factors. It has been seen this protein plays a key role in inflammatory reactions as the one's taking place in the tissue surrounding a tumor. This could explain why this protein gets suddenly overproduced in tumor cells. L1CAM's diverse functions make tumor cells more aggressive and resistant. Their migratory and motility related functions may result key in cell epithelial–mesenchymal transition (EMT) allowing cells to lose cell to cell static junctions and apico-basal polarity leading to them becoming migratory and independent. Also, its capacity to form adhesive interactions within different cell types may result in an advantage for tumor cells when it comes to co-opt and invade the surrounding tissues or capillaries.
Once tumor cells become anchorage-independent and migratory, due to L1 up-regulation, they leave the tissue where they belong and migrate through the capillaries to other organs. One frequent destination of tumor cells is the brain. So to settle in the brain, tumor cells have to succeed in crossing the blood brain barrier (BBB) where they get exposed to the plasmin secreted by astrocytes. Plasmin breaks L1CAM and inhibits the malignant cell's migrating powers. However, recent studies have noted these cancer cells overproduce anti-PA serpins, which are the usual inhibitors of plasmin, allowing them to cross the BBB and succeed in metastasis.[35]
Possible therapies involving L1CAM
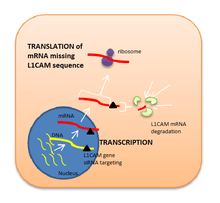
Because L1CAM is considered to be a key factor in metastasis, it has been suggested that blocking this protein may inhibit cancer cells migration and tumor progression. Antibody therapy directed against L1CAM in mice models of cancer block tumor growth but enhance EMT.[36] Liposome-encapsulated small interfering RNA has also proved to be an effective inhibitor for L1CAM expression as its function is to degrade a specific range of mRNA base pairs (in this case, the ones encoding for L1CAM sequence of amino acids) after transcription, so that the protein cannot be synthetised.[37] Nevertheless, these possible therapies involving L1CAM as a target in human cancer are still in preclinical research.[38]
References
- GRCh38: Ensembl release 89: ENSG00000198910 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000031391 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Samatov TR, Wicklein D, Tonevitsky AG (2016). "L1CAM: Cell adhesion and more". Progress in Histochemistry and Cytochemistry. 51 (2): 25–32. doi:10.1016/j.proghi.2016.05.001. PMID 27267927.
- "Entrez Gene: L1CAM L1 cell adhesion molecule".
- "L1CAM gene". Genetics Home Reference. U.S. Department of Health and Human Services.
- Djabali M, Mattei MG, Nguyen C, Roux D, Demengeot J, Denizot F, Moos M, Schachner M, Goridis C, Jordan BR (August 1990). "The gene encoding L1, a neural adhesion molecule of the immunoglobulin family, is located on the X chromosome in mouse and man". Genomics. 7 (4): 587–93. doi:10.1016/0888-7543(90)90203-7. PMID 2387585.
- "L1CAM Mutation Web Page". L1CAM Mutation Database. University Medical Center Groningen. 12 October 2012. Retrieved 23 October 2016.
- Bateman A, Jouet M, MacFarlane J, Du JS, Kenwrick S, Chothia C (1996). "Outline structure of the human L1 cell adhesion molecule and the sites where mutations cause neurological disorders". The EMBO Journal. 15 (22): 6050–9. doi:10.1002/j.1460-2075.1996.tb00993.x. PMC 452426. PMID 8947027.
- Hlavin ML, Lemmon V (1991). "Molecular structure and functional testing of human L1CAM: an interspecies comparison". Genomics. 11 (2): 416–23. doi:10.1016/0888-7543(91)90150-D. PMID 1769655.
- Haspel J, Grumet M (2003). "The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes". Frontiers in Bioscience. 8 (6): s1210–25. doi:10.2741/1108. PMID 12957823.
- http://atlasgeneticsoncology.org/Genes/L1CAMID44110chXq28.html
- Cushier A. "Cell interactions". University of Malta. Retrieved 2 October 2016.
- Becker WM, Kleinsmith LJ, Hardin J, Raasch J (2003). "Chapter 16: Cell-Cell Recognition and Adhesion, Cell Junctions". The World of the Cell (5th ed.). San Francisco: Benjamin/Cummings Pub. Co. pp. 302–14. ISBN 978-0-8053-4852-1.
- Moos M, Tacke R, Scherer H, Teplow D, Früh K, Schachner M (August 1988). "Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domains similar to fibronectin". Nature. 334 (6184): 701–3. Bibcode:1988Natur.334..701M. doi:10.1038/334701a0. PMID 3412448.
- Fransen E, Van Camp G, Vits L, Willems PJ (1997-01-01). "L1-associated diseases: clinical geneticists divide, molecular geneticists unite". Human Molecular Genetics. 6 (10): 1625–32. doi:10.1093/hmg/6.10.1625. PMID 9300653.
- Weller S, Gärtner J (2001-01-01). "Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): Mutations in the L1CAM gene". Human Mutation. 18 (1): 1–12. doi:10.1002/humu.1144. PMID 11438988.
- Kenwrick S, Watkins A, De Angelis E (April 2000). "Neural cell recognition molecule L1: relating biological complexity to human disease mutations". Human Molecular Genetics. 9 (6): 879–86. doi:10.1093/hmg/9.6.879. PMID 10767310.
- Schäfer MK, Altevogt P (July 2010). "L1CAM malfunction in the nervous system and human carcinomas". Cellular and Molecular Life Sciences. 67 (14): 2425–37. doi:10.1007/s00018-010-0339-1. PMID 20237819.
- Saugier-Veber P, Martin C, Le Meur N, Lyonnet S, Munnich A, David A, Hénocq A, Héron D, Jonveaux P, Odent S, Manouvrier S, Moncla A, Morichon N, Philip N, Satge D, Tosi M, Frébourg T (1998-01-01). "Identification of novel L1CAM mutations using fluorescence-assisted mismatch analysis". Human Mutation. 12 (4): 259–66. doi:10.1002/(SICI)1098-1004(1998)12:4<259::AID-HUMU7>3.0.CO;2-A. PMID 9744477.
- Zhang X, Davis JQ, Carpenter S, Bennett V (November 1998). "Structural requirements for association of neurofascin with ankyrin". The Journal of Biological Chemistry. 273 (46): 30785–94. doi:10.1074/jbc.273.46.30785. PMID 9804856.
- Garver TD, Ren Q, Tuvia S, Bennett V (May 1997). "Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin". The Journal of Cell Biology. 137 (3): 703–14. doi:10.1083/jcb.137.3.703. PMC 2139872. PMID 9151675.
- Bearer CF (October 2001). "Developmental neurotoxicity. Illustration of principles". Pediatric Clinics of North America. 48 (5): 1199–213, ix. doi:10.1016/s0031-3955(05)70369-2. PMID 11579669.
- Ikawa H, Kawano H, Takeda Y, Masuyama H, Watanabe K, Endo M, Yokoyama J, Kitajima M, Uyemura K, Kawamura K (April 1997). "Impaired expression of neural cell adhesion molecule L1 in the extrinsic nerve fibers in Hirschsprung's disease". Journal of Pediatric Surgery. 32 (4): 542–5. doi:10.1016/s0022-3468(97)90703-x. PMID 9126750.
- "L1CAM (L1 cell adhesion molecule)". Atlas of Genetics and Cytogenetics in Oncology and Hematology l. Retrieved 6 October 2016.
- "L1CAM Gene". Gene Cards.
- Reid RA, Hemperly JJ (1992). "Variants of human L1 cell adhesion molecule arise through alternate splicing of RNA". Journal of Molecular Neuroscience. 3 (3): 127–35. doi:10.1007/BF02919404. PMID 1627459.
- Pfeifer M, Schirmer U, Geismann C, Schäfer H, Sebens S, Altevogt P (2010). "L1CAM expression in endometrial carcinomas is regulated by usage of two different promoter regions". BMC Molecular Biology. 11: 64. doi:10.1186/1471-2199-11-64. PMC 2939505. PMID 20799950.
- Lund K, Dembinski JL, Solberg N, Urbanucci A, Mills IG, Krauss S (2015). "Slug-dependent upregulation of L1CAM is responsible for the increased invasion potential of pancreatic cancer cells following long-term 5-FU treatment". PLOS ONE. 10 (4): e0123684. Bibcode:2015PLoSO..1023684L. doi:10.1371/journal.pone.0123684. PMC 4393253. PMID 25860483.
- "L1CAM - Neural cell adhesion molecule L1 precursor - Homo sapiens (Human) - L1CAM gene & protein". www.uniprot.org. Retrieved 2016-10-23.
- Mikulak J, Negrini S, Klajn A, D'Alessandro R, Mavilio D, Meldolesi J (March 2012). "Dual REST-dependence of L1CAM: from gene expression to alternative splicing governed by Nova2 in neural cells". Journal of Neurochemistry. 120 (5): 699–709. doi:10.1111/j.1471-4159.2011.07626.x. PMID 22176577.
- "L1CAM - Neural cell adhesion molecule L1 precursor - Homo sapiens (Human) - L1CAM gene & protein". UniProt. Retrieved 2016-10-23.
- Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, Kaibuchi K (September 2003). "CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth". Nature Cell Biology. 5 (9): 819–26. doi:10.1038/ncb1039. PMID 12942088.
- Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, Massagué J (2014). "Serpins promote cancer cell survival and vascular co-option in brain metastasis". Cell. 156 (5): 1002–16. doi:10.1016/j.cell.2014.01.040. PMC 3988473. PMID 24581498.
- Doberstein K, Harter PN, Haberkorn U, Bretz NP, Arnold B, Carretero R, Moldenhauer G, Mittelbronn M, Altevogt P (March 2015). "Antibody therapy to human L1CAM in a transgenic mouse model blocks local tumor growth but induces EMT". International Journal of Cancer. 136 (5): E326–39. doi:10.1002/ijc.29222. PMID 25230579.
- Sung SY, Wu IH, Chuang PH, Petros JA, Wu HC, Zeng HJ, Huang WC, Chung LW, Hsieh CL (30 Oct 2014). "Targeting L1 cell adhesion molecule expression using liposome-encapsulated siRNA suppresses prostate cancer bone metastasis and growth". Oncotarget. 5 (20): 9911–29. doi:10.18632/oncotarget.2478. PMC 4259447. PMID 25294816.
- Altevogt P, Doberstein K, Fogel M (2016). "L1CAM in human cancer". International Journal of Cancer. 138 (7): 1565–76. doi:10.1002/ijc.29658. PMID 26111503.
Further reading
- Fransen E, Lemmon V, Van Camp G, Vits L, Coucke P, Willems PJ (1996). "CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1". European Journal of Human Genetics. 3 (5): 273–84. doi:10.1159/000472311. PMID 8556302.
- Bearer CF (October 2001). "L1 cell adhesion molecule signal cascades: targets for ethanol developmental neurotoxicity". Neurotoxicology. 22 (5): 625–33. doi:10.1016/S0161-813X(01)00034-1. PMID 11770884.
- Rosenthal A, Jouet M, Kenwrick S (October 1992). "Aberrant splicing of neural cell adhesion molecule L1 mRNA in a family with X-linked hydrocephalus". Nature Genetics. 2 (2): 107–12. doi:10.1038/ng1092-107. PMID 1303258.
- Fryns JP, Spaepen A, Cassiman JJ, van den Berghe H (June 1991). "X linked complicated spastic paraplegia, MASA syndrome, and X linked hydrocephalus owing to congenital stenosis of the aqueduct of Sylvius: variable expression of the same mutation at Xq28". Journal of Medical Genetics. 28 (6): 429–31. doi:10.1136/jmg.28.6.429-a. PMC 1016918. PMID 1870106.
- Rosenthal A, MacKinnon RN, Jones DS (October 1991). "PCR walking from microdissection clone M54 identifies three exons from the human gene for the neural cell adhesion molecule L1 (CAM-L1)". Nucleic Acids Research. 19 (19): 5395–401. doi:10.1093/nar/19.19.5395. PMC 328904. PMID 1923824.
- Kobayashi M, Miura M, Asou H, Uyemura K (October 1991). "Molecular cloning of cell adhesion molecule L1 from human nervous tissue: a comparison of the primary sequences of L1 molecules of different origin". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1090 (2): 238–40. doi:10.1016/0167-4781(91)90108-X. PMID 1932117.
- Harper JR, Prince JT, Healy PA, Stuart JK, Nauman SJ, Stallcup WB (March 1991). "Isolation and sequence of partial cDNA clones of human L1: homology of human and rodent L1 in the cytoplasmic region". Journal of Neurochemistry. 56 (3): 797–804. doi:10.1111/j.1471-4159.1991.tb01994.x. PMID 1993895.
- Wolff JM, Frank R, Mujoo K, Spiro RC, Reisfeld RA, Rathjen FG (August 1988). "A human brain glycoprotein related to the mouse cell adhesion molecule L1". The Journal of Biological Chemistry. 263 (24): 11943–7. PMID 3136168.
- Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M (May 1994). "The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth". The Journal of Cell Biology. 125 (3): 669–80. doi:10.1083/jcb.125.3.669. PMC 2119998. PMID 7513709.
- Ruiz JC, Cuppens H, Legius E, Fryns JP, Glover T, Marynen P, Cassiman JJ (July 1995). "Mutations in L1-CAM in two families with X linked complicated spastic paraplegia, MASA syndrome, and HSAS". Journal of Medical Genetics. 32 (7): 549–52. doi:10.1136/jmg.32.7.549. PMC 1050549. PMID 7562969.
- Olive S, Dubois C, Schachner M, Rougon G (November 1995). "The F3 neuronal glycosylphosphatidylinositol-linked molecule is localized to glycolipid-enriched membrane subdomains and interacts with L1 and fyn kinase in cerebellum". Journal of Neurochemistry. 65 (5): 2307–17. doi:10.1046/j.1471-4159.1995.65052307.x. PMID 7595520.
- Jouet M, Moncla A, Paterson J, McKeown C, Fryer A, Carpenter N, Holmberg E, Wadelius C, Kenwrick S (June 1995). "New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome". American Journal of Human Genetics. 56 (6): 1304–14. PMC 1801103. PMID 7762552.
- Fransen E, Schrander-Stumpel C, Vits L, Coucke P, Van Camp G, Willems PJ (December 1994). "X-linked hydrocephalus and MASA syndrome present in one family are due to a single missense mutation in exon 28 of the L1CAM gene". Human Molecular Genetics. 3 (12): 2255–6. doi:10.1093/hmg/3.12.2255. PMID 7881431.
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (July 1994). "X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene". Nature Genetics. 7 (3): 402–7. doi:10.1038/ng0794-402. PMID 7920659.
External links
- GeneReviews/NCBI/NIH/UW entry on L1 Syndrome
- L1+Cell+Adhesion+Molecule at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
Atlas of genetics and cytogenetics in oncology and haematology: http://atlasgeneticsoncology.org/Genes/L1CAMID44110chXq28.html