Exon
An exon is any part of a gene that will encode a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature messenger RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
History
The term exon derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons."[1]
This definition was originally made for protein-coding transcripts that are spliced before being translated. The term later came to include sequences removed from rRNA[2] and tRNA,[3] and it also was used later for RNA molecules originating from different parts of the genome that are then ligated by trans-splicing.[4]
Contribution to genomes and size distribution
Although unicellular eukaryotes such as yeast have either no introns or very few, metazoans and especially vertebrate genomes have a large fraction of non-coding DNA. For instance, in the human genome only 1.1% of the genome is spanned by exons, whereas 24% is in introns, with 75% of the genome being intergenic DNA.[5] This can provide a practical advantage in omics-aided health care (such as precision medicine) because it makes commercialized whole exome sequencing a smaller and less expensive challenge than commercialized whole genome sequencing. The large variation in genome size and C-value across life forms has posed an interesting challenge called the C-value enigma.
Across all eukaryotic genes in GenBank, there were (in 2002), on average, 5.48 exons per gene. The average exon encoded 30-36 amino acids.[6] While the longest exon in the human genome is 11555 bp long, several exons have been found to be only 2 bp long.[7] A single-nucleotide exon has been reported from the Arabidopsis genome.[8]
Structure and function

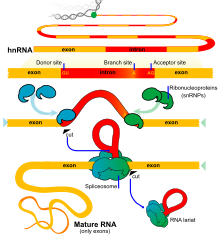
In protein-coding genes, the exons include both the protein-coding sequence and the 5′- and 3′-untranslated regions (UTR). Often the first exon includes both the 5′-UTR and the first part of the coding sequence, but exons containing only regions of 5′-UTR or (more rarely) 3′-UTR occur in some genes, i.e. the UTRs may contain introns.[9] Some non-coding RNA transcripts also have exons and introns.
Mature mRNAs originating from the same gene need not include the same exons, since different introns in the pre-mRNA can be removed by the process of alternative splicing.
Exonization is the creation of a new exon, as a result of mutations in introns.[10]
Experimental approaches using exons
Exon trapping or 'gene trapping' is a molecular biology technique that exploits the existence of the intron-exon splicing to find new genes.[11] The first exon of a 'trapped' gene splices into the exon that is contained in the insertional DNA. This new exon contains the ORF for a reporter gene that can now be expressed using the enhancers that control the target gene. A scientist knows that a new gene has been trapped when the reporter gene is expressed.
Splicing can be experimentally modified so that targeted exons are excluded from mature mRNA transcripts by blocking the access of splice-directing small nuclear ribonucleoprotein particles (snRNPs) to pre-mRNA using Morpholino antisense oligos.[12] This has become a standard technique in developmental biology. Morpholino oligos can also be targeted to prevent molecules that regulate splicing (e.g. splice enhancers, splice suppressors) from binding to pre-mRNA, altering patterns of splicing.
See also
References
- Gilbert W (February 1978). "Why genes in pieces?". Nature. 271 (5645): 501. doi:10.1038/271501a0. PMID 622185.
- Kister KP, Eckert WA (March 1987). "Characterization of an authentic intermediate in the self-splicing process of ribosomal precursor RNA in macronuclei of Tetrahymena thermophila". Nucleic Acids Research. 15 (5): 1905–20. doi:10.1093/nar/15.5.1905. PMC 340607. PMID 3645543.
- Valenzuela P, Venegas A, Weinberg F, Bishop R, Rutter WJ (January 1978). "Structure of yeast phenylalanine-tRNA genes: an intervening DNA segment within the region coding for the tRNA". Proceedings of the National Academy of Sciences of the United States of America. 75 (1): 190–4. doi:10.1073/pnas.75.1.190. PMC 411211. PMID 343104.
- Liu AY, Van der Ploeg LH, Rijsewijk FA, Borst P (June 1983). "The transposition unit of variant surface glycoprotein gene 118 of Trypanosoma brucei. Presence of repeated elements at its border and absence of promoter-associated sequences". Journal of Molecular Biology. 167 (1): 57–75. doi:10.1016/S0022-2836(83)80034-5. PMID 6306255.
- Venter J.C.; et al. (2000). "The Sequence of the Human Genome". Science. 291 (5507): 1304–51. doi:10.1126/science.1058040. PMID 11181995.
- Sakharkar M, Passetti F, de Souza JE, Long M, de Souza SJ (2002). "ExInt: an Exon Intron Database". Nucleic Acids Res. 30 (1): 191–4. doi:10.1093/nar/30.1.191. PMC 99089. PMID 11752290.
- Sakharkar M.K.; Chow VT; Kangueane P. (2004). "Distributions of exons and introns in the human genome". In Silico Biol. 4 (4): 387–93. PMID 15217358.
- Guo Lei, Liu Chun-Ming (2015). "A single-nucleotide exon found in Arabidopsis". Scientific Reports. 5: 18087. doi:10.1038/srep18087. PMC 4674806. PMID 26657562.
- Bicknell, AA (December 2012). "Introns in UTRs: Why we should stop ignoring them". BioEssays. 34 (12): 1025–1034. doi:10.1002/bies.201200073. PMID 23108796. S2CID 5808466.
- Sorek R (October 2007). "The birth of new exons: mechanisms and evolutionary consequences". RNA. 13 (10): 1603–8. doi:10.1261/rna.682507. PMC 1986822. PMID 17709368.
- Duyk G. M; Kim S. W.; Myers R. M; Cox D. R (1990). "Exon Trapping: a Genetic Screen to Identify Candidate Transcribed Sequences in Cloned Mammalian Genomic DNA". Proceedings of the National Academy of Sciences. 87 (22): 8995–8999. doi:10.1073/pnas.87.22.8995. PMC 55087. PMID 2247475.
- Morcos PA (June 2007). "Achieving targeted and quantifiable alteration of mRNA splicing with Morpholino oligos". Biochemical and Biophysical Research Communications. 358 (2): 521–7. doi:10.1016/j.bbrc.2007.04.172. PMID 17493584.
General references
- Zhang MQ (May 1998). "Statistical features of human exons and their flanking regions". Human Molecular Genetics. 7 (5): 919–32. doi:10.1093/hmg/7.5.919. PMID 9536098.
- Thanaraj TA, Robinson AJ (November 2000). "Prediction of exact boundaries of exons". Brief. Bioinform. 1 (4): 343–56. doi:10.1093/bib/1.4.343. PMID 11465052.