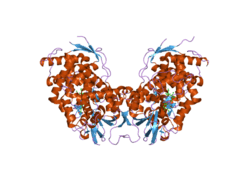
Cholesterol 7 alpha-hydroxylase
Cholesterol 7 alpha-hydroxylase also known as cholesterol 7-alpha-monooxygenase or cytochrome P450 7A1 (CYP7A1) is an enzyme that in humans is encoded by the CYP7A1 gene [5] which has an important role in cholesterol metabolism. It is a cytochrome P450 enzyme, which belongs to the oxidoreductase class, and converts cholesterol to 7-alpha-hydroxycholesterol, the first and rate limiting step in bile acid synthesis.
The inhibition of cholesterol 7-alpha-hydroxylase (CYP7A1) represses bile acid biosynthesis.[6]
Evolution
Sequence comparisons indicated a huge similarity between cytochromes P450 identified in man and bacteria, and suggested that the superfamily cytochrome P450 first originated from a common ancestral gene some three billion years ago.
The superfamily cytochrome P450 was named in 1961, because of the 450-nm spectral peak pigment that cytochrome P450 has when reduced and bound to carbon monoxide. In the early 1960s, P450 was thought to be one enzyme, and by the mid 1960s it was associated with drug and steroid metabolism.[7]
However, the membrane-associated and hydrophobic nature of the enzyme system impeded purification, and the number of proteins involved could not be accurately counted. Advances in mRNA purification in the early 1980s allowed to isolate the first cDNA encoding a complete cytochrome P450 (CYP) protein, and thereafter, results of many cloning studies have revealed a large number of different enzymes.[7]
Advances in molecular biology and genomics facilitated the biochemical characterisation of individual P450 enzymes:
- The cytochromes P450 act on many endogenous substrates, introducing oxidative, peroxidative, and reductive changes into small molecules of widely different chemical structures. Substrates identified to date include saturated and unsaturated fatty acids, eicosanoids, sterols and steroids, bile acids, vitamin D3 derivatives, retinoids, and uroporphyrinogens.[7]
- Many cytochrome P450 enzymes can metabolise various exogenous compounds including drugs, environmental chemicals and pollutants, and natural plant products.[7]
- Metabolism of foreign chemicals frequently results in successful detoxication of the irritant; However, the actions of P450 enzymes can also generate toxic metabolites that contribute to increased risks of cancer, birth defects, and other toxic effects.
- The expression of many P450 enzymes is often induced by accumulation of a substrate.
- The ability of one P450 substrate to affect the concentrations of another in this manner is the basis for so-called drug-drug interactions, which complicate treatment.[7]
Molecular structure
Cholesterol 7 alpha hydroxylase consists of 491 amino acids, which on folding forms 23 alpha helices and 26 beta sheets.[8][9]
Function
Cholesterol 7 alpha-hydroxylase is a cytochrome P450 heme enzyme that oxidizes cholesterol in the position 7 using molecular oxygen. It is an oxidoreductase. CYP7A1 is located in the endoplasmic reticulum (ER) and is important for the synthesis of bile acid and the regulation of cholesterol levels.[8][10]
Synthesis of bile acid
Cholesterol 7 alpha-hydroxylase is the rate-limiting enzyme in the synthesis of bile acid from cholesterol via the classic pathway, catalyzing the formation of 7α-hydroxycholesterol. The unique detergent properties of bile acids are essential for the digestion and intestinal absorption of hydrophobic nutrients.[8]
Bile acids have powerful toxic properties like membrane disruption and there are a wide range of mechanisms to restrict their accumulation in tissues and blood. The discovery of farnesoid X receptor (FXR) which is located in the liver, has opened new insights. Bile acid activation of FXR represses the expression of CYP7A1 via, raising the expression of small heterodimer (SHP), a non-DNA binding protein.[8]
The increased abundance of SHP causes it to associate with liver receptor homolog (LRH)-1, an obligate factor required for the transcription of CYP7A1. Furthermore, there is an "FXR/SHP-independent" mechanism that also represses CYP7A1 expression. This "FXR/SHP-independent" pathway involves the interaction of bile acids with liver macrophages, which finally induces the expression and secretion of cytokines. These inflammatory cytokines, which include tumor necrosis factor alpha and interleukin-1beta, act upon the liver parenchymal cells causing a rapid repression of the CYP7A1 gene.[8]
Regulation of activity
Regulation of CYP7A1 occurs at several levels including synthesis. Bile acids, steroid hormones, inflammatory cytokines, insulin, and growth factors inhibit CYP7A1 transcription through the 5′-upstream region of the promoter.[8] The average life of this enzyme is between two and three hours. Activity can be regulated by phosphorylation-dephosphorylation.
CYP7A1 is upregulated by the nuclear receptor LXR (liver X receptor) when cholesterol (to be specific, oxysterol) levels are high.[11] The effect of this upregulation is to increase the production of bile acids and reduce the level of cholesterol in hepatocytes.
It is downregulated by Sterol regulatory element-binding proteins (SREBP) when plasma cholesterol levels are low.
Bile acids provide feedback inhibition of CYP7A1 by at least two different pathways, both involving the farnesoid X receptor, FXR.[8] In the liver, bile acids bound to FXR induce small heterodimer partner, SHP which binds to LRH-1 and so inhibits the transcription of the enzyme. In the intestine, bile acids/FXR stimulate production of FGF15/19 (depending on species), which then acts as a hormone in the liver via FGFR4.[8]
Enzymatic mechanism
Specificity
One feature of enzymes is their high specificity. They are specific on a singular substrate, reaction or both together, that means, that the enzymes can catalyze all reactions wherein the substrate can experience.
The enzyme cholesterol 7 alpha hydroxylase catalyzes the reaction that converts cholesterol into cholesterol 7 alpha hydroxylase reducing and oxidizing that molecule.[8][12]
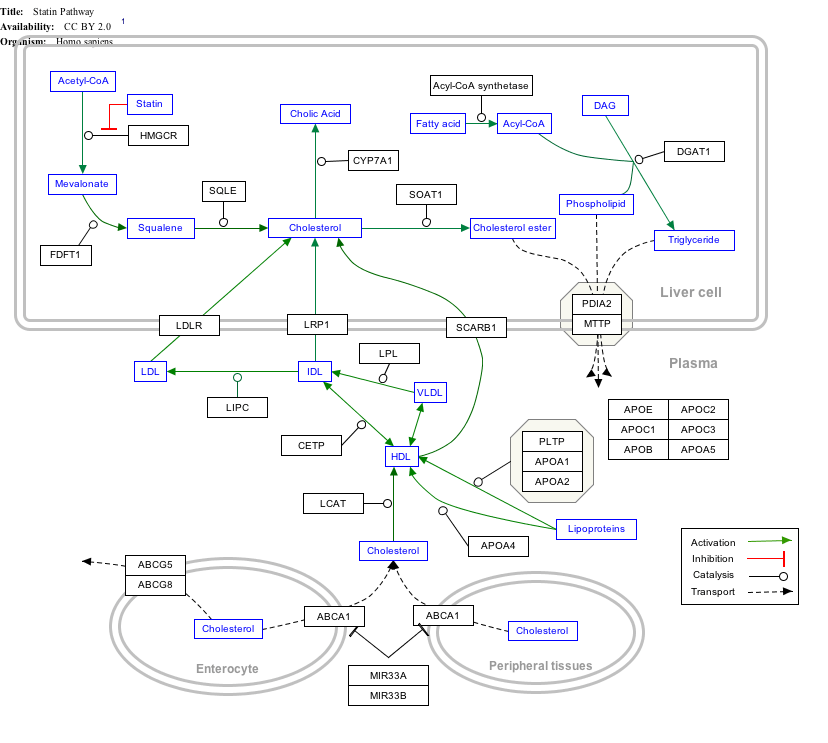
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
Clinical significance
Deficiency of this enzyme will increase the possibility of cholesterol gallstones.[13]
Disruption of CYP7A1 from classic bile acid synthesis in mice leads to either increased postnatal death or a milder phenotype with elevated serum cholesterol.[11] The latter is similar to the case in humans, where CYP7A1 mutations associate with high plasma low-density lipoprotein and hepatic cholesterol content, as well as deficient bile acid excretion. There is also a synergy between plasma low-density lipoprotein cholesterol (LDL-C) and risks of coronary artery disease (CAD).[11] Glucose signaling also induces CYP7A1 gene transcription by epigenetic regulation of the histone acetylation status. Glucose induction of bile acid synthesis have an important implication in metabolic control of glucose, lipid, and energy homeostasis under normal and diabetic conditions.[14] CYP7A1-rs3808607 and APOE isoform are associated with the extent of reduction in circulating LDL cholesterol in response to PS(define PS, Plant Sterol?) consumption and could serve as potential predictive genetic markers to identify individuals who would derive maximum LDL cholesterol lowering with PS consumption.[15] Genetic variations in CYP7A1 influence its expression and thus may affect the risk of gallstone disease and gallbladder cancer.[16]
One of the many lipid lowering effects of the fibrate drug class is mediated through the inhibition of transcription of this enzyme.[17] This inhibition leads to more cholesterol in the bile, which is the body's only route of cholesterol excretion. This also increases the risk of cholesterol gallstone formation.
Inhibition of CYP7A1 is thought to be involved in or responsible for the hepatotoxicity associated with ketoconazole.[18] The levorotatory enantiomer of ketoconazole, levoketoconazole, shows 12-fold reduced potency in inhibition of this enzyme, and is under development for certain indications (e.g., Cushing's syndrome) as a replacement for ketoconazole with reduced toxicity and improved tolerability and safety.[18]
See also
References
- GRCh38: Ensembl release 89: ENSG00000167910 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000028240 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Cohen JC, Cali JJ, Jelinek DF, Mehrabian M, Sparkes RS, Lusis AJ, Russell DW, Hobbs HH (Sep 1992). "Cloning of the human cholesterol 7 alpha-hydroxylase gene (CYP7) and localization to chromosome 8q11-q12". Genomics. 14 (1): 153–61. doi:10.1016/S0888-7543(05)80298-8. PMID 1358792.
- Miao J (2008). Regulation of Bile Acid Biosynthesis by Orphan Nuclear Receptor Small Heterodimer Partner (Ph.D.). University of Illinois at Urbana-Champaign.
- Nebert DW, Russell DW (2002). "Clinical importance of the cytochromes P450". Lancet. 360 (9340): 1155–62. doi:10.1016/S0140-6736(02)11203-7. PMID 12387968. S2CID 13577054.
- Chiang JY (October 2009). "Bile acids: regulation of synthesis". J. Lipid Res. 50 (10): 1955–66. doi:10.1194/jlr.R900010-JLR200. PMC 2739756. PMID 19346330.
- "RCSB PDB". RCSB PDB. Retrieved 2015-10-18.
- "Síntesis de Ácido Biliar, el Metabolismo y las Funciones Biológicas". Retrieved 2015-10-15.
- Chawla A, Saez E, Evans RM (Sep 2000). "Don't know much bile-ology". Cell. 103 (1): 1–4. doi:10.1016/S0092-8674(00)00097-0. PMID 11051540. S2CID 17408369.
- Hedstrom L (2010). "Enzyme Specificity and Selectivity". eLS Citable Reviews in the Life Sciences. doi:10.1002/9780470015902.a0000716.pub2. ISBN 978-0470016176.
- Paumgartner G, Sauerbruch T (Nov 1991). "Gallstones: pathogenesis". Lancet. 338 (8775): 1117–21. doi:10.1016/0140-6736(91)91972-W. PMID 1682550. S2CID 205037880.
- Li T, Chanda D, Zhang Y, Choi HS, Chiang JY (Apr 2010). "Glucose stimulates cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes". Journal of Lipid Research. 51 (4): 832–42. doi:10.1194/jlr.M002782. PMC 2842145. PMID 19965590.
- MacKay DS, Eck PK, Gebauer SK, Baer DJ, Jones PJ (Oct 2015). "CYP7A1-rs3808607 and APOE isoform associate with LDL cholesterol lowering after plant sterol consumption in a randomized clinical trial". The American Journal of Clinical Nutrition. 102 (4): 951–7. doi:10.3945/ajcn.115.109231. PMID 26333513.
- Srivastava A, Choudhuri G, Mittal B (2010). "CYP7A1 (-204 A>C; rs3808607 and -469 T>C; rs3824260) promoter polymorphisms and risk of gallbladder cancer in North Indian population". Metab. Clin. Exp. 59 (6): 767–73. doi:10.1016/j.metabol.2009.09.021. PMID 20005541.
- Gbaguidi GF, Agellon LB (2004-01-01). "The inhibition of the human cholesterol 7alpha-hydroxylase gene (CYP7A1) promoter by fibrates in cultured cells is mediated via the liver x receptor alpha and peroxisome proliferator-activated receptor alpha heterodimer". Nucleic Acids Research. 32 (3): 1113–21. doi:10.1093/nar/gkh260. PMC 373396. PMID 14960721.
- Cuevas-Ramos, Daniel; Lim, Dawn Shao Ting; Fleseriu, Maria (2016). "Update on medical treatment for Cushing's disease". Clinical Diabetes and Endocrinology. 2 (1): 16. doi:10.1186/s40842-016-0033-9. ISSN 2055-8260. PMC 5471955. PMID 28702250.
Further reading
- Davis RA, Miyake JH, Hui TY, Spann NJ (Apr 2002). "Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP". Journal of Lipid Research. 43 (4): 533–43. PMID 11907135.
- Kim HJ, Park HY, Kim E, Lee KS, Kim KK, Choi BO, Kim SM, Bae JS, Lee SO, Chun JY, Park TJ, Cheong HS, Jo I, Shin HD (Feb 2010). "Common CYP7A1 promoter polymorphism associated with risk of neuromyelitis optica". Neurobiology of Disease. 37 (2): 349–55. doi:10.1016/j.nbd.2009.10.013. PMID 19850125. S2CID 40067459.
- Holmes MV, Shah T, Vickery C, Smeeth L, Hingorani AD, Casas JP (2009). Luo Y (ed.). "Fulfilling the promise of personalized medicine? Systematic review and field synopsis of pharmacogenetic studies". PLOS ONE. 4 (12): e7960. Bibcode:2009PLoSO...4.7960H. doi:10.1371/journal.pone.0007960. PMC 2778625. PMID 19956635.
- Lipkin SM, Chao EC, Moreno V, Rozek LS, Rennert H, Pinchev M, Dizon D, Rennert G, Kopelovich L, Gruber SB (May 2010). "Genetic variation in 3-hydroxy-3-methylglutaryl CoA reductase modifies the chemopreventive activity of statins for colorectal cancer". Cancer Prevention Research. 3 (5): 597–603. doi:10.1158/1940-6207.CAPR-10-0007. PMID 20403997.
- Lambrinoudaki I, Kaparos G, Rizos D, Galapi F, Alexandrou A, Sergentanis TN, Creatsa M, Christodoulakos G, Kouskouni E, Botsis D (Aug 2009). "Apolipoprotein E and paraoxonase 1 polymorphisms are associated with lower serum thyroid hormones in postmenopausal women". Clinical Endocrinology. 71 (2): 284–90. doi:10.1111/j.1365-2265.2008.03476.x. PMID 19018779.
- Poduri A, Khullar M, Bahl A, Sharma YP, Talwar KK (Sep 2009). "A combination of proatherogenic single-nucleotide polymorphisms is associated with increased risk of coronary artery disease and myocardial infarction in Asian Indians". DNA and Cell Biology. 28 (9): 451–60. doi:10.1089/dna.2009.0887. PMID 19558216.
- Li T, Chanda D, Zhang Y, Choi HS, Chiang JY (Apr 2010). "Glucose stimulates cholesterol 7alpha-hydroxylase gene transcription in human hepatocytes". Journal of Lipid Research. 51 (4): 832–42. doi:10.1194/jlr.M002782. PMC 2842145. PMID 19965590.
- Kovár J, Lenícek M, Zimolová M, Vítek L, Jirsa M, Pitha J (2010). "Regulation of diurnal variation of cholesterol 7alpha-hydroxylase (CYP7A1) activity in healthy subjects". Physiological Research. 59 (2): 233–8. PMID 19537927.
- Chien KL, Wang KC, Chen YC, Chao CL, Hsu HC, Chen MF, Chen WJ (Mar 2010). "Common sequence variants in pharmacodynamic and pharmacokinetic pathway-related genes conferring LDL cholesterol response to statins". Pharmacogenomics. 11 (3): 309–17. doi:10.2217/pgs.09.160. PMID 20235787.
- Saito A, Kawamoto M, Kamatani N (Jun 2009). "Association study between single-nucleotide polymorphisms in 199 drug-related genes and commonly measured quantitative traits of 752 healthy Japanese subjects". Journal of Human Genetics. 54 (6): 317–23. doi:10.1038/jhg.2009.31. PMID 19343046.
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW (Jan 2004). "Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants". Pharmacogenetics. 14 (1): 1–18. doi:10.1097/00008571-200401000-00001. PMID 15128046. S2CID 18448751.
- Li T, Ma H, Park YJ, Lee YK, Strom S, Moore DD, Chiang JY (Oct 2009). "Forkhead box transcription factor O1 inhibits cholesterol 7alpha-hydroxylase in human hepatocytes and in high fat diet-fed mice". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1791 (10): 991–6. doi:10.1016/j.bbalip.2009.05.004. PMC 2743791. PMID 19463968.
- Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, Banerjee-Basu S, Baron-Cohen S (Jun 2009). "Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger syndrome". Autism Research. 2 (3): 157–77. doi:10.1002/aur.80. PMID 19598235.
- Barcelos AL, Chies R, Almeida SE, Fiegenbaum M, Schweigert ID, Chula FG, Rossetti ML, Silva CM (Jun 2009). "Association of CYP7A1 -278A>C polymorphism and the response of plasma triglyceride after dietary intervention in dyslipidemic patients". Brazilian Journal of Medical and Biological Research. 42 (6): 487–93. doi:10.1590/s0100-879x2009000600003. PMID 19448895.
- Voora D, Shah SH, Reed CR, Zhai J, Crosslin DR, Messer C, Salisbury BA, Ginsburg GS (Dec 2008). "Pharmacogenetic predictors of statin-mediated low-density lipoprotein cholesterol reduction and dose response". Circulation: Cardiovascular Genetics. 1 (2): 100–6. doi:10.1161/CIRCGENETICS.108.795013. PMC 2995295. PMID 20031551.
- Schwarz M, Lund EG, Russell DW (Apr 1998). "Two 7 alpha-hydroxylase enzymes in bile acid biosynthesis". Current Opinion in Lipidology. 9 (2): 113–8. doi:10.1097/00041433-199804000-00006. PMID 9559267.
- Beigneux A, Hofmann AF, Young SG (Jul 2002). "Human CYP7A1 deficiency: progress and enigmas". The Journal of Clinical Investigation. 110 (1): 29–31. doi:10.1172/JCI16076. PMC 151039. PMID 12093884.
- Ruaño G, Bernene J, Windemuth A, Bower B, Wencker D, Seip RL, Kocherla M, Holford TR, Petit WA, Hanks S (Feb 2009). "Physiogenomic comparison of edema and BMI in patients receiving rosiglitazone or pioglitazone". Clinica Chimica Acta; International Journal of Clinical Chemistry. 400 (1–2): 48–55. doi:10.1016/j.cca.2008.10.009. PMID 18996102.
- Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL (Apr 2009). "High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis". Hepatology. 49 (4): 1228–35. doi:10.1002/hep.22771. PMID 19185005.
- Srivastava A, Choudhuri G, Mittal B (Jun 2010). "CYP7A1 (-204 A>C; rs3808607 and -469 T>C; rs3824260) promoter polymorphisms and risk of gallbladder cancer in North Indian population". Metabolism. 59 (6): 767–73. doi:10.1016/j.metabol.2009.09.021. PMID 20005541.
External links
- Cholesterol+7-alpha-Hydroxylase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P22680 (Cytochrome P450 7A1) at the PDBe-KB.