Bile acid
Bile acids are steroid acids found predominantly in the bile of mammals and other vertebrates. Diverse bile acids are synthesized in the liver.[1] Bile acids are conjugated with taurine or glycine residues to give anions called bile salts.[2][3][4]
Primary bile acids are those synthesized by the liver. Secondary bile acids result from bacterial actions in the colon. In humans, taurocholic acid and glycocholic acid (derivatives of cholic acid) and taurochenodeoxycholic acid and glycochenodeoxycholic acid (derivatives of chenodeoxycholic acid) are the major bile salts. They are roughly equal in concentration.[5] The salts of their 7-alpha-dehydroxylated derivatives, deoxycholic acid and lithocholic acid, are also found, with derivatives of cholic, chenodeoxycholic and deoxycholic acids accounting for over 90% of human biliary bile acids.[5]
Bile acids comprise about 80% of the organic compounds in bile (others are phospholipids and cholesterol).[5] An increased secretion of bile acids produces an increase in bile flow. Bile acids facilitate digestion of dietary fats and oils. They serve as micelle-forming surfactants, which encapsulate nutrients, facilitating their absorption.[6] These micelles are suspended in the chyme before further processing. Bile acids also have hormonal actions throughout the body, particularly through the farnesoid X receptor and GPBAR1 (also known as TGR5).[7]
Production
Bile acid synthesis occurs in liver cells, which synthesize primary bile acids (cholic acid and chenodeoxycholic acid in humans) via cytochrome P450-mediated oxidation of cholesterol in a multi-step process. Approximately 600 mg of bile salts are synthesized daily to replace bile acids lost in the feces, although, as described below, much larger amounts are secreted, reabsorbed in the gut and recycled. The rate-limiting step in synthesis is the addition of a hydroxyl group of the 7th position of the steroid nucleus by the enzyme cholesterol 7 alpha-hydroxylase. This enzyme is down-regulated by cholic acid, up-regulated by cholesterol and is inhibited by the actions of the ileal hormone FGF15/19.[2][3]
Prior to secreting any of the bile acids (primary or secondary, see below), liver cells conjugate them with either glycine or taurine, to form a total of 8 possible conjugated bile acids. These conjugated bile acids are often referred to as bile salts. The pKa of the unconjugated bile acids are between 5 and 6.5,[4] and the pH of the duodenum ranges between 3 and 5, so when unconjugated bile acids are in the duodenum, they are almost always protonated (HA form), which makes them relatively insoluble in water. Conjugating bile acids with amino acids lowers the pKa of the bile-acid/amino-acid conjugate to between 1 and 4. Thus conjugated bile acids are almost always in their deprotonated (A-) form in the duodenum, which makes them much more water-soluble and much more able to fulfil their physiologic function of emulsifying fats.[8][9]
Once secreted into the lumen of the intestine, bile salts are modified by gut bacteria. They are partial dehydroxylated. Their glycine and taurine groups are removed to give the secondary bile acids, deoxycholic acid and lithocholic acid. Cholic acid is converted into deoxycholic acid and chenodeoxycholic acid into lithocholic acid. All four of these bile acids recycled, in a process known as enterohepatic circulation.[2][3]
Functions
As amphipathic molecules with hydrophobic and hydrophilic regions, conjugated bile salts sit at the lipid/water interface and, at the right concentration, form micelles.[9] The added solubility of conjugated bile salts aids in their function by preventing passive re-absorption in the small intestine. As a result, the concentration of bile acids/salts in the small intestine is high enough to form micelles and solubilize lipids. "Critical micellar concentration" refers to both an intrinsic property of the bile acid itself and amount of bile acid necessary to function in the spontaneous and dynamic formation of micelles.[9] Bile acid-containing micelles aid lipases to digest lipids and bring them near the intestinal brush border membrane, which results in fat absorption.[6]
Synthesis of bile acids is a major route of cholesterol metabolism in most species other than humans. The body produces about 800 mg of cholesterol per day and about half of that is used for bile acid synthesis producing 400–600 mg daily. Human adults secrete between 12-18 g of bile acids into the intestine each day, mostly after meals. The bile acid pool size is between 4–6 g, which means that bile acids are recycled several times each day. About 95% of bile acids are reabsorbed by active transport in the ileum and recycled back to the liver for further secretion into the biliary system and gallbladder. This enterohepatic circulation of bile acids allows a low rate of synthesis, only about 0.3g/day, but with large amounts being secreted into the intestine.[5]
Bile acids have other functions, including eliminating cholesterol from the body, driving the flow of bile to eliminate certain catabolites (including bilirubin), emulsifying fat-soluble vitamins to enable their absorption, and aiding in motility and the reduction of the bacteria flora found in the small intestine and biliary tract.[5]
Bile acids have metabolic actions in the body resembling those of hormones, acting through two specific receptors, the farnesoid X receptor and G protein-coupled bile acid receptor/TGR5.[7][10] They bind less specifically to some other receptors and have been reported to regulate the activity of certain enzymes [11] and ion channels [12] and the synthesis of diverse substances including endogenous fatty acid ethanolamides.[13] [14]
Structure and synthesis
- The structures of the principal human bile acids

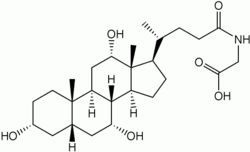

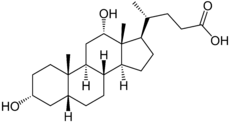
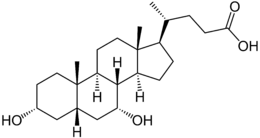
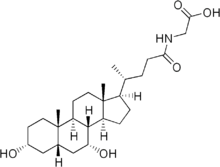
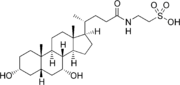
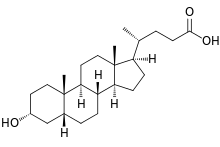
Bile salts constitute a large family of molecules, composed of a steroid structure with four rings, a five- or eight-carbon side-chain terminating in a carboxylic acid, and several hydroxyl groups, the number and orientation of which is different among the specific bile salts.[1] The four rings are labeled A, B, C, and D, from the farthest to the closest to the side chain with the carboxyl group. The D-ring is smaller by one carbon than the other three. The structure is commonly drawn with A at the left and D at the right. The hydroxyl groups can be in either of two configurations: either up (or out), termed beta (β; often drawn by convention as a solid line), or down, termed alpha (α; displayed as a dashed line). All bile acids have a 3-hydroxyl group, derived from the parent molecule, cholesterol, in which the 3-hydroxyl is beta.[1]
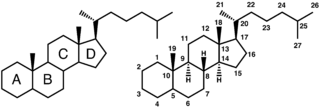
The initial step in the classical pathway of hepatic synthesis of bile acids is the enzymatic addition of a 7α hydroxyl group by cholesterol 7α-hydroxylase (CYP7A1) forming 7α-hydroxycholesterol. This is then metabolised to 7α-hydroxy-4-cholesten-3-one. There are multiple steps in bile acid synthesis requiring 14 enzymes in all.[3] These result in the junction between the first two steroid rings (A and B) being altered, making the molecule bent; in this process, the 3-hydroxyl is converted to the α orientation. The simplest 24-carbon bile acid has two hydroxyl groups at positions 3α and 7α. This is 3α,7α-dihydroxy-5β-cholan-24-oic acid, or, as more usually known, chenodeoxycholic acid. This bile acid was first isolated from the domestic goose, from which the "cheno" portion of the name was derived. The 5β in the name denotes the orientation of the junction between rings A and B of the steroid nucleus (in this case, they are bent). The term "cholan" denotes a particular steroid structure of 24 carbons, and the "24-oic acid" indicates that the carboxylic acid is found at position 24, at the end of the side-chain. Chenodeoxycholic acid is made by many species, and is the prototypic functional bile acid.[2][3]
An alternative (acidic) pathway of bile acid synthesis is initiated by mitochondrial sterol 27-hydroxylase (CYP27A1), expressed in liver, and also in macrophages and other tissues. CYP27A1 contributes significantly to total bile acid synthesis by catalyzing sterol side chain oxidation, after which cleavage of a three-carbon unit in the peroxisomes leads to formation of a C24 bile acid. Minor pathways initiated by 25-hydroxylase in the liver and 24-hydroxylase in the brain also may contribute to bile acid synthesis. 7α-hydroxylase (CYP7B1) generates oxysterols, which may be further converted in the liver to CDCA.[2][3]
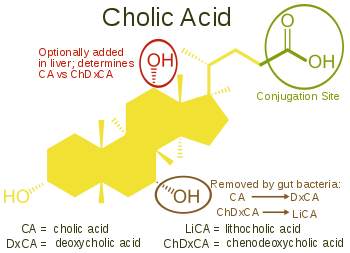
Cholic acid, 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid, the most abundant bile acid in humans and many other species, was discovered before chenodeoxycholic acid. It is a tri-hydroxy-bile acid with 3 hydroxyl groups (3α, 7α and 12α). In its synthesis in the liver, 12α hydroxylation is performed by the additional action of CYP8B1. As this had already been described, the discovery of chenodeoxcholic acid (with 2 hydroxyl groups) made this new bile acid a "deoxycholic acid" in that it had one fewer hydroxyl group than cholic acid.[2][3]
Deoxycholic acid is formed from cholic acid by 7-dehydroxylation, resulting in 2 hydroxyl groups (3α and 12α). This process with chenodeoxycholic acid results in a bile acid with only a 3α hydroxyl group, termed lithocholic acid (litho = stone) having been identified first in a gallstone from a calf. It is poorly water-soluble and rather toxic to cells.[2][3]
Different vertebrate families have evolved to use modifications of most positions on the steroid nucleus and side-chain of the bile acid structure. To avoid the problems associated with the production of lithocholic acid, most species add a third hydroxyl group to chenodeoxycholic acid. The subsequent removal of the 7α hydroxyl group by intestinal bacteria will then result in a less toxic but still-functional dihydroxy bile acid. Over the course of vertebrate evolution, a number of positions have been chosen for placement of the third hydroxyl group. Initially, the 16α position was favored, in particular in birds. Later, this position was superseded in a large number of species selecting the 12α position. Primates (including humans) utilize 12α for their third hydroxyl group position, producing cholic acid. In mice and other rodents, 6β hydroxylation forms muricholic acids (α or β depending on the 7 hydroxyl position). Pigs have 6α hydroxylation in hyocholic acid (3α,6α,7α-trihydroxy-5β-cholanoic acid), and other species have a hydroxyl group on position 23 of the side-chain.
Ursodeoxycholic acid was first isolated from bear bile, which has been used medicinally for centuries. Its structure resembles chenodeoxycholic acid but with the 7-hydroxyl group in the β position.[1]
Obeticholic acid, 6α-ethyl-chenodeoxycholic acid, is a semi-synthetic bile acid with greater activity as FXR agonist which is undergoing investigation as a pharmaceutical agent.
Hormonal actions
Bile acids also act as steroid hormones, secreted from the liver, absorbed from the intestine and having various direct metabolic actions in the body through the nuclear receptor Farnesoid X receptor (FXR), also known by its gene name NR1H4.[15][16][17] Another bile acid receptor is the cell membrane receptor known as G protein-coupled bile acid receptor 1 or TGR5. Many of their functions as signaling molecules in the liver and the intestines are by activating FXR, whereas TGR5 may be involved in metabolic, endocrine and neurological functions.[7]
Regulation of synthesis
As surfactants or detergents, bile acids are potentially toxic to cells, and so their concentrations are tightly regulated. Activation of FXR in the liver inhibits synthesis of bile acids, and is one mechanism of feedback control when bile acid levels are too high. Secondly, FXR activation by bile acids during absorption in the intestine increases transcription and synthesis of FGF19, which then inhibits bile acid synthesis in the liver.[18]
Metabolic functions
Emerging evidence associates FXR activation with alterations in triglyceride metabolism, glucose metabolism, and liver growth.[7][19]
Other interactions
Bile acids bind to some other proteins in addition to their hormone receptors (FXR and TGR5) and their transporters. Among these protein targets, the enzyme N-acyl phosphatidylethanolamine-specific phospholipase D (NAPE-PLD) generates bioactive lipid amides (e.g. the endogenous cannabinoid anandamide) that play important roles in several physiological pathways including stress and pain responses, appetite, and lifespan. NAPE-PLD orchestrates a direct cross-talk between lipid amide signals and bile acid physiology.[13]
Clinical significance
Hyperlipidemia
As bile acids are made from endogenous cholesterol, disruption of the enterohepatic circulation of bile acids will lower cholesterol. Bile acid sequestrants bind bile acids in the gut, preventing reabsorption. In so doing, more endogenous cholesterol is shunted into the production of bile acids, thereby lowering cholesterol levels. The sequestered bile acids are then excreted in the feces.[20]
Cholestasis
Tests for bile acids are useful in both human and veterinary medicine, as they aid in the diagnosis of a number of conditions, including types of cholestasis such as intrahepatic cholestasis of pregnancy, portosystemic shunt, and hepatic microvascular dysplasia in dogs.[21] Structural or functional abnormalities of the biliary system result in an increase in bilirubin (jaundice) and in bile acids in the blood. Bile acids are related to the itching (pruritus) which is common in cholestatic conditions such as primary biliary cirrhosis (PBC), primary sclerosing cholangitis or intrahepatic cholestasis of pregnancy.[22] Treatment with ursodeoxycholic acid has been used for many years in these cholestatic disorders.[23][24]
Gallstones
The relationship of bile acids to cholesterol saturation in bile and cholesterol precipitation to produce gallstones has been studied extensively. Gallstones may result from increased saturation of cholesterol or bilirubin, or from bile stasis. Lower concentrations of bile acids or phospholipids in bile reduce cholesterol solubility and lead to microcrystal formation. Oral therapy with chenodeoxycholic acid and/or ursodeoxycholic acid has been used to dissolve cholesterol gallstones.[25][26][27] Stones may recur when treatment is stopped. Bile acid therapy may be of value to prevent stones in certain circumstances such as following bariatric surgery.[28]
Bile acid diarrhea
Excess concentrations of bile acids in the colon are a cause of chronic diarrhea. It is commonly found when the ileum is abnormal or has been surgically removed, as in Crohn's disease, or cause a condition that resembles diarrhea-predominant irritable bowel syndrome (IBS-D). This condition of bile acid diarrhea/bile acid malabsorption can be diagnosed by the SeHCAT test and treated with bile acid sequestrants.[29]
Bile acids and colon cancer
Bile acids may have some importance in the development of colorectal cancer.[30] Deoxycholic acid (DCA) is increased in the colonic contents of humans in response to a high fat diet.[31] In populations with a high incidence of colorectal cancer, fecal concentrations of bile acids are higher,[32][33] and this association suggests that increased colonic exposure to bile acids could play a role in the development of cancer. In one particular comparison, the fecal DCA concentrations in Native Africans in South Africa (who eat a low fat diet) compared to African Americans (who eat a higher fat diet) was 7.30 vs. 37.51 nmol/g wet weight stool.[34] Native Africans in South Africa have a low incidence rate of colon cancer of less than 1:100,000,[35] compared to the high incidence rate for male African Americans of 72:100,000.[36]
Experimental studies also suggest mechanisms for bile acids in colon cancer. Exposure of colonic cells to high DCA concentrations increase formation of reactive oxygen species, causing oxidative stress, and also increase DNA damage.[37] Mice fed a diet with added DCA mimicking colonic DCA levels in humans on a high fat diet developed colonic neoplasia, including adenomas and adenocarcinomas (cancers), unlike mice fed a control diet producing one-tenth the level of colonic DCA who had no colonic neoplasia.[38][39]
The effects of ursodeoxycholic acid (UDCA) in modifying the risk of colorectal cancer has been looked at in several studies, particularly in primary sclerosing cholangitis and inflammatory bowel disease, with varying results partly related to dosage.[40][41] Genetic variation in the key bile acid synthesis enzyme, CYP7A1, influenced the effectiveness of UDCA in colorectal adenoma prevention in a large trial.[42]
Dermatology
Bile acids may be used in subcutaneous injections to remove unwanted fat (see Mesotherapy). Deoxycholic acid as an injectable has received FDA approval to dissolve submental fat.[43] Phase III trials showed significant responses although many subjects had mild adverse reactions of bruising, swelling, pain, numbness, erythema, and firmness around the treated area.[44][45]
References
- Hofmann AF, Hagey LR, Krasowski MD (February 2010). "Bile salts of vertebrates: structural variation and possible evolutionary significance". J. Lipid Res. 51 (2): 226–46. doi:10.1194/jlr.R000042. PMC 2803226. PMID 19638645.
- Russell DW (2003). "The enzymes, regulation, and genetics of bile acid synthesis". Annu. Rev. Biochem. 72: 137–74. doi:10.1146/annurev.biochem.72.121801.161712. PMID 12543708.
- Chiang JY (October 2009). "Bile acids: regulation of synthesis". J. Lipid Res. 50 (10): 1955–66. doi:10.1194/jlr.R900010-JLR200. PMC 2739756. PMID 19346330.
- Carey, MC.; Small, DM. (Oct 1972). "Micelle formation by bile salts. Physical-chemical and thermodynamic considerations". Arch Intern Med. 130 (4): 506–27. doi:10.1001/archinte.1972.03650040040005. PMID 4562149.
- Hofmann AF (1999). "The continuing importance of bile acids in liver and intestinal disease". Arch. Intern. Med. 159 (22): 2647–58. doi:10.1001/archinte.159.22.2647. PMID 10597755.
- Hofmann AF, Borgström B (February 1964). "The intraluminal phase of fat digestion in man: the lipid content of the micellar and oil phases of intestinal content obtained during fat digestion and absorption". J. Clin. Invest. 43 (2): 247–57. doi:10.1172/JCI104909. PMC 289518. PMID 14162533.
- Fiorucci S, Mencarelli A, Palladino G, Cipriani S (November 2009). "Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders". Trends Pharmacol. Sci. 30 (11): 570–80. doi:10.1016/j.tips.2009.08.001. PMID 19758712.
- 'Essentials of Medical Biochemistry, Lieberman, Marks and Smith, eds, p432, 2007'
- Hofmann AF (October 1963). "The function of bile salts in fat absorption. The solvent properties of dilute micellar solutions of conjugated bile salts". Biochem. J. 89: 57–68. doi:10.1042/bj0890057. PMC 1202272. PMID 14097367.
- Li T, Chiang JY (2014). "Bile acid signaling in metabolic disease and drug therapy". Pharmacol. Rev. 66 (4): 948–83. doi:10.1124/pr.113.008201. PMC 4180336. PMID 25073467.
- Nagahashi M, Takabe K, Liu R, Peng K, Wang X, Wang Y, Hait NC, Wang X, Allegood JC, Yamada A, Aoyagi T, Liang J, Pandak WM, Spiegel S, Hylemon PB, Zhou H (2015). "Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression". Hepatology. 61 (4): 1216–26. doi:10.1002/hep.27592. PMC 4376566. PMID 25363242.
- Wiemuth D, Sahin H, Falkenburger BH, Lefevre CM, Wasmuth HE, Grunder S (2012). "BASIC--a bile acid-sensitive ion channel highly expressed in bile ducts". FASEB J. 26 (10): 4122–30. doi:10.1096/fj.12-207043. PMID 22735174.
- Magotti P, Bauer I, Igarashi M, Babagoli M, Marotta R, Piomelli D, Garau G (2015). "Structure of Human N-Acylphosphatidylethanolamine-Hydrolyzing Phospholipase D: Regulation of Fatty Acid Ethanolamide Biosynthesis by Bile Acids". Structure. 23 (3): 598–604. doi:10.1016/j.str.2014.12.018. PMC 4351732. PMID 25684574.
- Margheritis, E, Castellani B, Magotti P, Peruzzi S, Romeo E, Natali F, Mostarda S, Gioiello A, Piomelli D, Garau G (2016). "Bile Acid Recognition by NAPE-PLD". ACS Chemical Biology. 11 (10): 2908–2914. doi:10.1021/acschembio.6b00624. PMC 5074845. PMID 27571266.
- Makishima M, Okamoto AY, Repa JJ, et al. (May 1999). "Identification of a nuclear receptor for bile acids". Science. 284 (5418): 1362–5. Bibcode:1999Sci...284.1362M. doi:10.1126/science.284.5418.1362. PMID 10334992.
- Parks DJ, Blanchard SG, Bledsoe RK, et al. (May 1999). "Bile acids: natural ligands for an orphan nuclear receptor". Science. 284 (5418): 1365–8. Bibcode:1999Sci...284.1365P. doi:10.1126/science.284.5418.1365. PMID 10334993.
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM (May 1999). "Endogenous bile acids are ligands for the nuclear receptor FXR/BAR". Mol. Cell. 3 (5): 543–53. doi:10.1016/s1097-2765(00)80348-2. PMID 10360171.
- Kim, I; Ahn, SH; Inagaki, T; Choi, M; Ito, S; Guo, GL; Kliewer, SA; Gonzalez, FJ (2007). "Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine". Journal of Lipid Research. 48 (12): 2664–72. doi:10.1194/jlr.M700330-JLR200. PMID 17720959.
- Shapiro, Hagit; Kolodziejczyk, Aleksandra A.; Halstuch, Daniel; Elinav, Eran (2018-01-16). "Bile acids in glucose metabolism in health and disease". Journal of Experimental Medicine. 215 (2): 383–396. doi:10.1084/jem.20171965. ISSN 0022-1007. PMC 5789421. PMID 29339445.
- Davidson MH (2011). "A systematic review of bile acid sequestrant therapy in children with familial hypercholesterolemia". J Clin Lipidol. 5 (2): 76–81. doi:10.1016/j.jacl.2011.01.005. PMID 21392720.
- Allen L, Stobie D, Mauldin GN, Baer KE (January 1999). "Clinicopathologic features of dogs with hepatic microvascular dysplasia with and without portosystemic shunts: 42 cases (1991-1996)". J. Am. Vet. Med. Assoc. 214 (2): 218–20. PMID 9926012.
- Pusl T, Beuers U (2007). "Intrahepatic cholestasis of pregnancy". Orphanet J Rare Dis. 2: 26. doi:10.1186/1750-1172-2-26. PMC 1891276. PMID 17535422.
- Poupon RE, Balkau B, Eschwège E, Poupon R (May 1991). "A multicenter, controlled trial of ursodiol for the treatment of primary biliary cirrhosis. UDCA-PBC Study Group". N. Engl. J. Med. 324 (22): 1548–54. doi:10.1056/NEJM199105303242204. PMID 1674105.
- Glantz A, Marschall HU, Lammert F, Mattsson LA (December 2005). "Intrahepatic cholestasis of pregnancy: a randomized controlled trial comparing dexamethasone and ursodeoxycholic acid". Hepatology. 42 (6): 1399–405. doi:10.1002/hep.20952. PMID 16317669.
- Danzinger RG, Hofmann AF, Schoenfield LJ, Thistle JL (January 1972). "Dissolution of cholesterol gallstones by chenodeoxycholic acid". N. Engl. J. Med. 286 (1): 1–8. doi:10.1056/NEJM197201062860101. PMID 5006919.
- Thistle JL, Hofmann AF (September 1973). "Efficacy and specificity of chenodeoxycholic acid therapy for dissolving gallstones". N. Engl. J. Med. 289 (13): 655–9. doi:10.1056/NEJM197309272891303. PMID 4580472.
- Petroni ML, Jazrawi RP, Pazzi P, et al. (January 2001). "Ursodeoxycholic acid alone or with chenodeoxycholic acid for dissolution of cholesterol gallstones: a randomized multicentre trial. The British-Italian Gallstone Study group". Aliment. Pharmacol. Ther. 15 (1): 123–8. doi:10.1046/j.1365-2036.2001.00853.x. PMID 11136285.
- Uy MC, Talingdan-Te MC, Espinosa WZ, Daez ML, Ong JP (December 2008). "Ursodeoxycholic acid in the prevention of gallstone formation after bariatric surgery: a meta-analysis". Obes Surg. 18 (12): 1532–8. doi:10.1007/s11695-008-9587-7. PMID 18574646.
- Pattni, S; Walters, JR (2009). "Recent advances in the understanding of bile acid malabsorption". British Medical Bulletin. 92: 79–93. doi:10.1093/bmb/ldp032. PMID 19900947.
- Degirolamo C, Modica S, Palasciano G, Moschetta A (2011). "Bile acids and colon cancer: Solving the puzzle with nuclear receptors". Trends Mol Med. 17 (10): 564–72. doi:10.1016/j.molmed.2011.05.010. PMID 21724466.
- Reddy BS, Hanson D, Mangat S, et al. (September 1980). "Effect of high-fat, high-beef diet and of mode of cooking of beef in the diet on fecal bacterial enzymes and fecal bile acids and neutral sterols". J. Nutr. 110 (9): 1880–7. doi:10.1093/jn/110.9.1880. PMID 7411244.
- Hill MJ (May 1990). "Bile flow and colon cancer". Mutat. Res. 238 (3): 313–20. doi:10.1016/0165-1110(90)90023-5. PMID 2188127.
- Cheah PY (1990). "Hypotheses for the etiology of colorectal cancer--an overview". Nutr Cancer. 14 (1): 5–13. doi:10.1080/01635589009514073. PMID 2195469.
- Ou J, DeLany JP, Zhang M, Sharma S, O'Keefe SJ (2012). "Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations". Nutr Cancer. 64 (1): 34–40. doi:10.1080/01635581.2012.630164. PMC 6844083. PMID 22136517.
- O'Keefe SJ, Kidd M, Espitalier-Noel G, Owira P (May 1999). "Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber". Am. J. Gastroenterol. 94 (5): 1373–80. PMID 10235221.
- American Cancer Society. Cancer Facts and Figures 2009. http://www.cancer.org/Research/CancerFactsFigures/cancer-facts-figures-2009
- Bernstein H, Bernstein C, Payne CM, Dvorak K (July 2009). "Bile acids as endogenous etiologic agents in gastrointestinal cancer". World J. Gastroenterol. 15 (27): 3329–40. doi:10.3748/wjg.15.3329. PMC 2712893. PMID 19610133.
- Bernstein C, Holubec H, Bhattacharyya AK, et al. (August 2011). "Carcinogenicity of deoxycholate, a secondary bile acid". Arch. Toxicol. 85 (8): 863–71. doi:10.1007/s00204-011-0648-7. PMC 3149672. PMID 21267546.
- Prasad AR, Prasad S, Nguyen H, Facista A, Lewis C, Zaitlin B, Bernstein H, Bernstein C (Jul 2014). "Novel diet-related mouse model of colon cancer parallels human colon cancer". World J Gastrointest Oncol. 6 (7): 225–43. doi:10.4251/wjgo.v6.i7.225. PMC 4092339. PMID 25024814.CS1 maint: multiple names: authors list (link)
- Singh S, Khanna S, Pardi DS, Loftus EV, Talwalkar JA (2013). "Effect of ursodeoxycholic acid use on the risk of colorectal neoplasia in patients with primary sclerosing cholangitis and inflammatory bowel disease: a systematic review and meta-analysis". Inflamm. Bowel Dis. 19 (8): 1631–8. doi:10.1097/MIB.0b013e318286fa61. PMID 23665966.
- Eaton JE, Silveira MG, Pardi DS, Sinakos E, Kowdley KV, Luketic VA, Harrison ME, McCashland T, Befeler AS, Harnois D, Jorgensen R, Petz J, Lindor KD (2011). "High-dose ursodeoxycholic acid is associated with the development of colorectal neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis". Am. J. Gastroenterol. 106 (9): 1638–45. doi:10.1038/ajg.2011.156. PMC 3168684. PMID 21556038.
- Wertheim BC, Smith JW, Fang C, Alberts DS, Lance P, Thompson PA (2012). "Risk modification of colorectal adenoma by CYP7A1 polymorphisms and the role of bile acid metabolism in carcinogenesis". Cancer Prev Res (Phila). 5 (2): 197–204. doi:10.1158/1940-6207.CAPR-11-0320. PMC 3400261. PMID 22058145.
- "Deoxycholic acid injection". Medline plus. Retrieved 26 August 2015.
- Ascher B, Hoffmann K, Walker P, Lippert S, Wollina U, Havlickova B (2014). "Efficacy, patient-reported outcomes and safety profile of ATX-101 (deoxycholic acid), an injectable drug for the reduction of unwanted submental fat: results from a phase III, randomized, placebo-controlled study". J Eur Acad Dermatol Venereol. 28 (12): 1707–15. doi:10.1111/jdv.12377. PMC 4263247. PMID 24605812.
- Wollina U, Goldman A (2015). "ATX-101 for reduction of submental fat". Expert Opin Pharmacother. 16 (5): 755–62. doi:10.1517/14656566.2015.1019465. PMID 25724831.
External links
- Bile+Acids+and+Salts at the US National Library of Medicine Medical Subject Headings (MeSH)
- Special Issue on "Bile Acids"