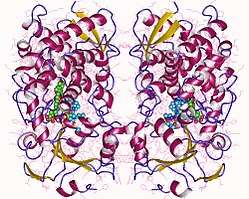
CYP3A4
Cytochrome P450 3A4 (abbreviated CYP3A4) (EC 1.14.13.97) is an important enzyme in the body, mainly found in the liver and in the intestine. It oxidizes small foreign organic molecules (xenobiotics), such as toxins or drugs, so that they can be removed from the body.
While many drugs are deactivated by CYP3A4, there are also some drugs which are activated by the enzyme. Some substances, such as grapefruit juice and some drugs, interfere with the action of CYP3A4. These substances will therefore either amplify or weaken the action of those drugs that are modified by CYP3A4.
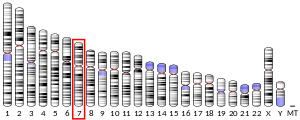
CYP3A4 is a member of the cytochrome P450 family of oxidizing enzymes. Several other members of this family are also involved in drug metabolism, but CYP3A4 is the most common and the most versatile one. Like all members of this family, it is a hemoprotein, i.e. a protein containing a heme group with an iron atom. In humans, the CYP3A4 protein is encoded by the CYP3A4 gene.[3] This gene is part of a cluster of cytochrome P450 genes on chromosome 7q22.1.[4]
Function
CYP3A4 is a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases that catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids, and other lipids components.
The CYP3A4 protein localizes to the endoplasmic reticulum, and its expression is induced by glucocorticoids and some pharmacological agents. Cytochrome P450 enzymes metabolize approximately 60% of prescribed drugs, with CYP3A4 responsible for about half of this metabolism;[5] substrates include acetaminophen, codeine, ciclosporin (cyclosporin), diazepam, and erythromycin. The enzyme also metabolizes some steroids and carcinogens.[6] Most drugs undergo deactivation by CYP3A4, either directly or by facilitated excretion from the body. Also, many substances are bioactivated by CYP3A4 to form their active compounds, and many protoxins being toxicated into their toxic forms (for examples – see table below).
CYP3A4 also possesses epoxygenase activity in that it metabolizes arachidonic acid to epoxyeicosatrienoic acids (EETs), i.e. (±)-8,9-, (±)-11,12-, and (±)-14,15-epoxyeicosatrienoic acids.[7] The EETs have a wide range of activities including the promotion of certain types of cancers (see epoxyeicosatetraenoic acid#cancer). CYP3A4 promotes the growth of various types of human cancer cell lines in culture by producing (±)-14,15-epoxyeicosatrienoic acids which stimulate these cells to grow.[8] The cytochrome P450 is also reported to have fatty acid monooxgenase activity for metabolizing arachidonic acid to 20-Hydroxyeicosatetraenoic acid (20-HETE).[9] 20-HETE has a wide range of activities that also include growth stimulation in breast and other types of cancers (see 12-hydroxyeicosatetraenoic acid#cancer).
Evolution
The CYP3A4 gene exhibits a much more complicated upstream regulatory region in comparison with its paralogs.[10] This increased complexity renders the CYP3A4 gene more sensitive to endogenous and exogenous PXR and CAR ligands, instead of relying on gene variants for wider specificity.[10] Chimpanzee and human CYP3A4 are highly conserved in metabolism of many ligands, although four amino acids positively selected in humans led to a 5-fold benzylation of 7-BFC in the presence of the hepatotoxic secondary bile acid lithocholic acid.[11] This change in consequence contributes to an increased human defense against cholestasis.[11]
Tissue distribution
Fetuses tend to not express CYP3A4 in their liver tissue, but rather CYP3A7 (EC 1.14.14.1), which acts on a similar range of substrates. CYP3A4 is absent in fetal liver but increases to approximately 40% of adult levels in the fourth month of life and 72% at 12 months.[12][13]
Although CYP3A4 is predominantly found in the liver, it is also present in other organs and tissues of the body, where it may play an important role in metabolism. CYP3A4 in the intestine plays an important role in the metabolism of certain drugs. Often this allows prodrugs to be activated and absorbed – as in the case of the histamine H1-receptor antagonist terfenadine.
Recently CYP3A4 has also been identified in the brain, however its role in the central nervous system is still unknown.[14]
Mechanisms
Cytochrome P450 enzymes perform an assortment of modifications on a variety of ligands, utilizing its large active site and its ability to bind more than one substrate at a time to perform complicated chemical alterations in the metabolism of endogenous and exogenous compounds. These include hydroxylation, epoxidation of olefins, aromatic oxidation, heteroatom oxidations, N- and O- dealkylation reactions, aldehyde oxidations, dehydrogenation reactions, and aromatase activity.[15][16]
Hydroxylation of an sp3 C-H bond is one of the ways in which CYP3A4 (and cytochrome P450 oxygenases) affects its ligand.[17] In fact, hydroxylation is sometimes followed by dehydrogenation, leading to more complex metabolites.[16] An example of a molecule that undergoes more than one reaction due to CYP3A4 includes tamoxifen, which is hydroxylated to 4-hydroxy-tamoxifen and then dehydrated to 4-hydroxy-tamoxifen quinone methide.[16] Two mechanisms have been proposed as the primary pathway of hydroxylation in P450 enzymes.
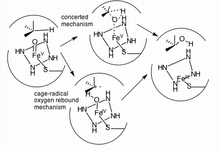
The first pathway suggested is a cage-controlled radical method ("oxygen rebound"), and the second involves a concerted mechanism that does not utilize a radical intermediate but instead acts very quickly via a "radical clock".[17]
Inhibition through fruit ingestion
In 1998, various researchers showed that grapefruit juice, and grapefruit in general, is a potent inhibitor of CYP3A4, which can affect the metabolism of a variety of drugs, increasing their bioavailability.[18][19][20][21][22] In some cases, this can lead to a fatal interaction with drugs like astemizole or terfenadine.[19] The effect of grapefruit juice with regard to drug absorption was originally discovered in 1989. The first published report on grapefruit drug interactions was in 1991 in the Lancet entitled "Interactions of Citrus Juices with Felodipine and Nifedipine", and was the first reported food-drug interaction clinically. The effects of grapefruit last from 3–7 days, with the greatest effects when juice is taken an hour previous to administration of the drug.[23]
In addition to grapefruit, other fruits have similar effects. Noni (M. citrifolia), for example, is a dietary supplement typically consumed as a juice and also inhibits CYP3A4;[24] pomegranate juice has this effect as well.[25]
Variability
While over 28 single nucleotide polymorphisms (SNPs) have been identified in the CYP3A4 gene, it has been found that this does not translate into significant interindividual variability in vivo. It can be supposed that this may be due to the induction of CYP3A4 on exposure to substrates.
CYP3A4 alleles which have been reported to have minimal function compared to wild-type include CYP3A4*6 (an A17776 insertion) and CYP3A4*17 (F189S). Both of these SNPs led to decreased catalytic activity with certain ligands, including testosterone and nifedipine in comparison to wild-type metabolism.[26]
Variability in CYP3A4 function can be determined noninvasively by the erythromycin breath test (ERMBT). The ERMBT estimates in vivo CYP3A4 activity by measuring the radiolabelled carbon dioxide exhaled after an intravenous dose of (14C-N-methyl)-erythromycin.[27]
Induction
CYP3A4 is induced by a wide variety of ligands. These ligands bind to the pregnane X receptor (PXR). The activated PXR complex forms a heterodimer with the retinoid X receptor (RXR), which binds to the XREM region of the CYP3A4 gene. XREM is a regulatory region of the CYP3A4 gene, and binding causes a cooperative interaction with proximal promoter regions of the gene, resulting in increased transcription and expression of CYP3A4. Activation of the PXR/RXR heterodimer initiates transcription of the CYP3A4 promoter region and gene. Ligand binding increases when in the presence of CYP3A4 ligands, such as in the presence of aflatoxin B1, M1, and G1. Indeed, due to the enzyme's large and malleable active site, it is possible for the enzyme to bind multiple ligands at once, leading to potentially detrimental side effects.[28]
Induction of CYP3A4 has been shown to vary in humans depending on sex. Evidence shows an increased drug clearance by CYP3A4 in women, even when accounting for differences in body weight. A study by Wolbold et al. (2003) found that the median CYP3A4 levels measured from surgically removed liver samples of a random sample of women exceeded CYP3A4 levels in the livers of men by 129%. CYP3A4 mRNA transcripts were found in similar proportions, suggesting a pre-translational mechanism for the up-regulation of CYP3A4 in women. The exact cause of this elevated level of enzyme in women is still under speculation, however studies have elucidated other mechanisms (such as CYP3A5 or CYP3A7 compensation for lowered levels of CYP3A4) that affect drug clearance in both men and women.[29]
CYP3A4 substrate activation varies amongst different animal species. Certain ligands activate human PXR, which promotes CYP3A4 transcription, while showing no activation in other species. For instance, mouse PXR is not activated by rifampicin and human PXR is not activated by pregnenalone 16α-carbonitrile[30] In order to facilitate study of CYP3A4 functional pathways in vivo, mouse strains have been developed using transgenes in order to produce null/human CYP3A4 and PXR crosses. Although humanized hCYP3A4 mice successfully expressed the enzyme in their intestinal tract, low levels of hCYP3A4 were found in the liver.[30] This effect has been attributed to CYP3A4 regulation by the growth hormone signal transduction pathway.[30] In addition to providing an in vivo model, humanized CYP3A4 mice (hCYP3A4) have been used to further emphasize gender differences in CYP3A4 activity.[30]
CYP3A4 activity levels have also been linked to diet and environmental factors, such as duration of exposure to xenobiotic substances.[31] Due to the enzyme's extensive presence in the intestinal mucosa, the enzyme has shown sensitivity to starvation symptoms and is upregulated in defense of adverse effects. Indeed, in fatheaded minnows, unfed female fish were shown to have increased PXR and CYP3A4 expression, and displayed a more pronounced response to xenobiotic factors after exposure after several days of starvation.[31] By studying animal models and keeping in mind the innate differences in CYP3A4 activation, investigators can better predict drug metabolism and side effects in human CYP3A4 pathways.
Turnover
Estimates of the turnover rate of human CYP3A4 vary widely. For hepatic CYP3A4, in vivo methods yield estimates of enzyme half-life mainly in the range of 70 to 140 hours, whereas in vitro methods give estimates from 26 to 79 hours.[32] Turnover of gut CYP3A4 is likely to be a function of the rate of enterocyte renewal; an indirect approach based on recovery of activity following exposure to grapefruit juice yields measurements in the 12- to 33-hour range.[32]
Technology
Due to membrane-bound CYP3A4's natural propensity to conglomerate, it has historically been difficult to study drug binding in both solution and on surfaces. Co-crystallization is difficult since the substrates tend to have a low Kd (between 5-150 μM) and low solubility in aqueous solutions.[33] A successful strategy in isolating the bound enzyme is the functional stabilization of monomeric CYP3A4 on silver nanoparticles produced from nanosphere lithography and analyzed via localized surface plasmon resonance spectroscopy (LSPR).[34] These analyses can be used as a high-sensitivity assay of drug binding, and may become integral in further high-throughput assays utilized in initial drug discovery testing. In addition to LSPR, CYP3A4-Nanodisc complexes have been found helpful in other applications including solid-state NMR, redox potentiometry, and steady-state enzyme kinetics.[34]
Ligands
Following is a table of selected substrates, inducers and inhibitors of CYP3A4. Where classes of agents are listed, there may be exceptions within the class.
Inhibitors of CYP3A4 can be classified by their potency, such as:
- Strong inhibitor being one that causes at least a 5-fold increase in the plasma AUC values, or more than 80% decrease in clearance.[35]
- Moderate inhibitor being one that causes at least a 2-fold increase in the plasma AUC values, or 50-80% decrease in clearance.[35]
- Weak inhibitor being one that causes at least a 1.25-fold but less than 2-fold increase in the plasma AUC values, or 20-50% decrease in clearance.[35]
| Substrates | Inhibitors | Inducers |
|---|---|---|
|
Strong
Moderate
Weak
Unspecified potency
|
Strong potency Unspecified potency
|
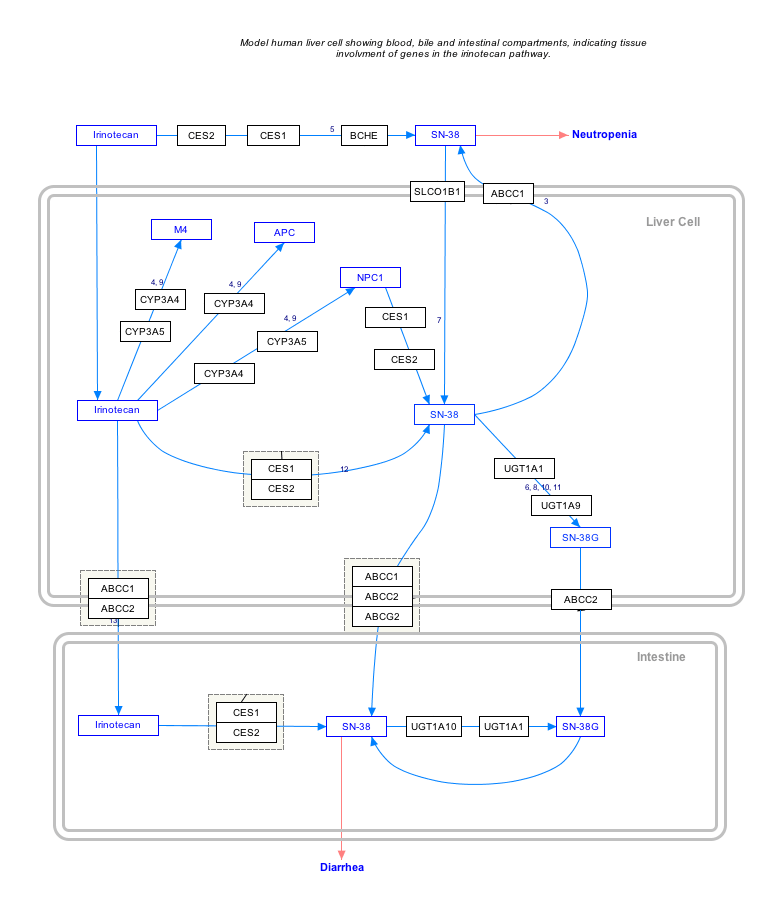
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- The interactive pathway map can be edited at WikiPathways: "IrinotecanPathway_WP46359".
See also
- List of drugs affected by grapefruit
References
- GRCh38: Ensembl release 89: ENSG00000160868 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hashimoto H, Toide K, Kitamura R, Fujita M, Tagawa S, Itoh S, Kamataki T (December 1993). "Gene structure of CYP3A4, an adult-specific form of cytochrome P450 in human livers, and its transcriptional control". European Journal of Biochemistry / FEBS. 218 (2): 585–95. doi:10.1111/j.1432-1033.1993.tb18412.x. PMID 8269949.
- Inoue K, Inazawa J, Nakagawa H, Shimada T, Yamazaki H, Guengerich FP, Abe T (June 1992). "Assignment of the human cytochrome P-450 nifedipine oxidase gene (CYP3A4) to chromosome 7 at band q22.1 by fluorescence in situ hybridization". The Japanese Journal of Human Genetics. 37 (2): 133–8. doi:10.1007/BF01899734. PMID 1391968.
- Zanger UM, Schwab M (April 2013). "Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation". Pharmacology & Therapeutics. 138 (1): 103–41. doi:10.1016/j.pharmthera.2012.12.007. PMID 23333322.
- EntrezGene 1576
- Bishop-Bailey D, Thomson S, Askari A, Faulkner A, Wheeler-Jones C (2014). "Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism". Annual Review of Nutrition. 34: 261–79. doi:10.1146/annurev-nutr-071813-105747. PMID 24819323.
- Fleming I (October 2014). "The pharmacology of the cytochrome P450 epoxygenase/soluble epoxide hydrolase axis in the vasculature and cardiovascular disease". Pharmacological Reviews. 66 (4): 1106–40. doi:10.1124/pr.113.007781. PMID 25244930.
- Miyata N, Taniguchi K, Seki T, Ishimoto T, Sato-Watanabe M, Yasuda Y, Doi M, Kametani S, Tomishima Y, Ueki T, Sato M, Kameo K (June 2001). "HET0016, a potent and selective inhibitor of 20-HETE synthesizing enzyme". British Journal of Pharmacology. 133 (3): 325–9. doi:10.1038/sj.bjp.0704101. PMC 1572803. PMID 11375247.
- Qiu H, Mathäs M, Nestler S, Bengel C, Nem D, Gödtel-Armbrust U, Lang T, Taudien S, Burk O, Wojnowski L (March 2010). "The unique complexity of the CYP3A4 upstream region suggests a nongenetic explanation of its expression variability". Pharmacogenetics and Genomics. 20 (3): 167–78. doi:10.1097/FPC.0b013e328336bbeb. PMID 20147837. S2CID 205602787.
- Kumar S, Qiu H, Oezguen N, Herlyn H, Halpert JR, Wojnowski L (June 2009). "Ligand diversity of human and chimpanzee CYP3A4: activation of human CYP3A4 by lithocholic acid results from positive selection". Drug Metabolism and Disposition. 37 (6): 1328–33. doi:10.1124/dmd.108.024372. PMC 2683693. PMID 19299527.
- Johnson TN, Rostami-Hodjegan A, Tucker GT (2006). "Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children". Clinical Pharmacokinetics. 45 (9): 931–56. doi:10.2165/00003088-200645090-00005. PMID 16928154. S2CID 25596506.
- Johnson TN, Tucker GT, Rostami-Hodjegan A (May 2008). "Development of CYP2D6 and CYP3A4 in the first year of life". Clinical Pharmacology and Therapeutics. 83 (5): 670–1. doi:10.1038/sj.clpt.6100327. PMID 18043691.
- Robertson GR, Field J, Goodwin B, Bierach S, Tran M, Lehnert A, Liddle C (July 2003). "Transgenic mouse models of human CYP3A4 gene regulation". Molecular Pharmacology. 64 (1): 42–50. doi:10.1124/mol.64.1.42. PMID 12815159. S2CID 17209434.
- Schmiedlin-Ren P, Edwards DJ, Fitzsimmons ME, He K, Lown KS, Woster PM, Rahman A, Thummel KE, Fisher JM, Hollenberg PF, Watkins PB (November 1997). "Mechanisms of enhanced oral availability of CYP3A4 substrates by grapefruit constituents. Decreased enterocyte CYP3A4 concentration and mechanism-based inactivation by furanocoumarins". Drug Metabolism and Disposition. 25 (11): 1228–33. PMID 9351897.
- Shahrokh K, Cheatham TE, Yost GS (October 2012). "Conformational dynamics of CYP3A4 demonstrate the important role of Arg212 coupled with the opening of ingress, egress and solvent channels to dehydrogenation of 4-hydroxy-tamoxifen". Biochimica et Biophysica Acta (BBA) - General Subjects. 1820 (10): 1605–17. doi:10.1016/j.bbagen.2012.05.011. PMC 3404218. PMID 22677141.
- Meunier B, de Visser SP, Shaik S (September 2004). "Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes". Chemical Reviews. 104 (9): 3947–80. doi:10.1021/cr020443g. PMID 15352783. S2CID 33927145.
- He K, Iyer KR, Hayes RN, Sinz MW, Woolf TF, Hollenberg PF (April 1998). "Inactivation of cytochrome P450 3A4 by bergamottin, a component of grapefruit juice". Chemical Research in Toxicology. 11 (4): 252–9. doi:10.1021/tx970192k. PMID 9548795.
- Bailey DG, Malcolm J, Arnold O, Spence JD (August 1998). "Grapefruit juice-drug interactions". British Journal of Clinical Pharmacology. 46 (2): 101–10. doi:10.1046/j.1365-2125.1998.00764.x. PMC 1873672. PMID 9723817.
- Garg SK, Kumar N, Bhargava VK, Prabhakar SK (September 1998). "Effect of grapefruit juice on carbamazepine bioavailability in patients with epilepsy". Clinical Pharmacology and Therapeutics. 64 (3): 286–8. doi:10.1016/S0009-9236(98)90177-1. PMID 9757152.
- Bailey DG, Dresser GK (2004). "Interactions between grapefruit juice and cardiovascular drugs". American Journal of Cardiovascular Drugs. 4 (5): 281–97. doi:10.2165/00129784-200404050-00002. PMID 15449971. S2CID 11525439.
- Bressler R (November 2006). "Grapefruit juice and drug interactions. Exploring mechanisms of this interaction and potential toxicity for certain drugs". Geriatrics. 61 (11): 12–8. PMID 17112309.
- Lilja JJ, Kivistö KT, Neuvonen PJ (October 2000). "Duration of effect of grapefruit juice on the pharmacokinetics of the CYP3A4 substrate simvastatin". Clinical Pharmacology and Therapeutics. 68 (4): 384–90. doi:10.1067/mcp.2000.110216. PMID 11061578.
- "Integrative Medicine, Noni". Memorial Sloan-Kettering Cancer Center. Retrieved 2013-06-27.
- Hidaka M, Okumura M, Fujita K, Ogikubo T, Yamasaki K, Iwakiri T, Setoguchi N, Arimori K (May 2005). "Effects of pomegranate juice on human cytochrome p450 3A (CYP3A) and carbamazepine pharmacokinetics in rats". Drug Metabolism and Disposition. 33 (5): 644–8. doi:10.1124/dmd.104.002824. PMID 15673597. S2CID 7997718.
- Lee SJ, Goldstein JA (June 2005). "Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests". Pharmacogenomics. 6 (4): 357–71. doi:10.1517/14622416.6.4.357. PMID 16004554.
- Watkins PB (August 1994). "Noninvasive tests of CYP3A enzymes". Pharmacogenetics. 4 (4): 171–84. doi:10.1097/00008571-199408000-00001. PMID 7987401.
- Ratajewski M, Walczak-Drzewiecka A, Sałkowska A, Dastych J (August 2011). "Aflatoxins upregulate CYP3A4 mRNA expression in a process that involves the PXR transcription factor". Toxicology Letters. 205 (2): 146–53. doi:10.1016/j.toxlet.2011.05.1034. PMID 21641981.
- Wolbold R, Klein K, Burk O, Nüssler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM (October 2003). "Sex is a major determinant of CYP3A4 expression in human liver". Hepatology. 38 (4): 978–88. doi:10.1053/jhep.2003.50393. PMID 14512885.
- Gonzalez FJ (2007). "CYP3A4 and pregnane X receptor humanized mice". Journal of Biochemical and Molecular Toxicology. 21 (4): 158–62. doi:10.1002/jbt.20173. PMID 17936928.
- Crago J, Klaper RD (September 2011). "Influence of gender, feeding regimen, and exposure duration on gene expression associated with xenobiotic metabolism in fathead minnows (Pimephales promelas)". Comparative Biochemistry and Physiology. Toxicology & Pharmacology. 154 (3): 208–12. doi:10.1016/j.cbpc.2011.05.016. PMID 21664292.
- Yang J, Liao M, Shou M, Jamei M, Yeo KR, Tucker GT, Rostami-Hodjegan A (June 2008). "Cytochrome p450 turnover: regulation of synthesis and degradation, methods for determining rates, and implications for the prediction of drug interactions". Current Drug Metabolism. 9 (5): 384–94. doi:10.2174/138920008784746382. PMID 18537575.
- Sevrioukova IF, Poulos TL (January 2012). "Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand". The Journal of Biological Chemistry. 287 (5): 3510–7. doi:10.1074/jbc.M111.317081. PMC 3271004. PMID 22157006.
- Das A, Zhao J, Schatz GC, Sligar SG, Van Duyne RP (May 2009). "Screening of type I and II drug binding to human cytochrome P450-3A4 in nanodiscs by localized surface plasmon resonance spectroscopy". Analytical Chemistry. 81 (10): 3754–9. doi:10.1021/ac802612z. PMC 4757437. PMID 19364136.
- Flockhart DA (2007). "Drug Interactions: Cytochrome P450 Drug Interaction Table". Indiana University School of Medicine. Retrieved on December 25, 2008.
- Where classes of agents are listed, there may be exceptions within the class
- FASS (drug formulary): Swedish environmental classification of pharmaceuticals Facts for prescribers (Fakta för förskrivare). Retrieved July 2011
- "Erlotinib".
Metabolized primarily by CYP3A4 and, to a lesser degree, by CYP1A2 and the extrahepatic isoform CYP1A1
- "Cyclobenzaprine". DrugBank.
- Moody DE, Fang WB, Lin SN, Weyant DM, Strom SC, Omiecinski CJ (December 2009). "Effect of rifampin and nelfinavir on the metabolism of methadone and buprenorphine in primary cultures of human hepatocytes". Drug Metabolism and Disposition. 37 (12): 2323–9. doi:10.1124/dmd.109.028605. PMC 2784702. PMID 19773542.
- Hutchinson MR, Menelaou A, Foster DJ, Coller JK, Somogyi AA (Mar 2004). "CYP2D6 and CYP3A4 involvement in the primary oxidative metabolism of hydrocodone by human liver microsomes". British Journal of Clinical Pharmacology. 57 (3): 287–97. doi:10.1046/j.1365-2125.2003.02002.x. PMC 1884456. PMID 14998425.
- Tanaka, E. (October 1999). "Clinically significant pharmacokinetic drug interactions with benzodiazepines". Journal of Clinical Pharmacy and Therapeutics. 24 (5): 347–355. doi:10.1046/j.1365-2710.1999.00247.x. ISSN 0269-4727. PMID 10583697.
- Sutton D, Butler AM, Nadin L, Murray M (July 1997). "Role of CYP3A4 in human hepatic diltiazem N-demethylation: inhibition of CYP3A4 activity by oxidized diltiazem metabolites". The Journal of Pharmacology and Experimental Therapeutics. 282 (1): 294–300. PMID 9223567.
- "Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers". U S Food and Drug Administration Home Page. 2009-06-25. Retrieved 2019-02-01.
- Lown KS, Bailey DG, Fontana RJ, Janardan SK, Adair CH, Fortlage LA, Brown MB, Guo W, Watkins PB (May 1997). "Grapefruit juice increases felodipine oral availability in humans by decreasing intestinal CYP3A protein expression". The Journal of Clinical Investigation. American Society for Clinical Investigation. 99 (10): 2545–53. doi:10.1172/jci119439. PMC 508096. PMID 9153299.
- Bailey DG, Bend JR, Arnold JM, Tran LT, Spence JD (July 1996). "Erythromycin-felodipine interaction: magnitude, mechanism, and comparison with grapefruit juice". Clinical Pharmacology and Therapeutics. Springer Nature. 60 (1): 25–33. doi:10.1016/s0009-9236(96)90163-0. PMID 8689808.
- Guengerich FP, Brian WR, Iwasaki M, Sari MA, Bäärnhielm C, Berntsson P (June 1991). "Oxidation of dihydropyridine calcium channel blockers and analogues by human liver cytochrome P-450 IIIA4". Journal of Medicinal Chemistry. 34 (6): 1838–44. doi:10.1021/jm00110a012. PMID 2061924.
- Katoh M, Nakajima M, Yamazaki H, Yokoi T (February 2001). "Inhibitory effects of CYP3A4 substrates and their metabolites on P-glycoprotein-mediated transport". European Journal of Pharmaceutical Sciences. 12 (4): 505–13. doi:10.1016/s0928-0987(00)00215-3. PMID 11231118.
- Foti RS, Rock DA, Wienkers LC, Wahlstrom JL (June 2010). "Selection of alternative CYP3A4 probe substrates for clinical drug interaction studies using in vitro data and in vivo simulation". Drug Metabolism and Disposition. American Society for Pharmacology & Experimental Therapeutics (ASPET). 38 (6): 981–7. doi:10.1124/dmd.110.032094. PMID 20203109. S2CID 6823063.
- Odou P, Ferrari N, Barthélémy C, Brique S, Lhermitte M, Vincent A, Libersa C, Robert H (April 2005). "Grapefruit juice-nifedipine interaction: possible involvement of several mechanisms". Journal of Clinical Pharmacy and Therapeutics. 30 (2): 153–8. doi:10.1111/j.1365-2710.2004.00618.x. PMID 15811168.
- "NIFEDIPINE EXTENDED RELEASE- nifedipine tablet, extended release". DailyMed. 2012-11-29. Retrieved 2019-02-01.
Drug Interactions: Nifedipine is mainly eliminated by metabolism and is a substrate of CYP3A. Inhibitors and inducers of CYP3A can impact the exposure to nifedipine and consequently its desirable and undesirable effects. In vitro and in vivo data indicate that nifedipine can inhibit the metabolism of drugs that are substrates of CYP3A, thereby increasing the exposure to other drugs. Nifedipine is a vasodilator, and coadministration of other drugs affecting blood pressure may result in pharmacodynamic interactions.
- Zhang Y, Guo X, Lin ET, Benet LZ (April 1998). "Overlapping substrate specificities of cytochrome P450 3A and P-glycoprotein for a novel cysteine protease inhibitor". Drug Metabolism and Disposition. 26 (4): 360–6. PMID 9531525.
- Stringer KA, Mallet J, Clarke M, Lindenfeld JA (1992). "The effect of three different oral doses of verapamil on the disposition of theophylline". European Journal of Clinical Pharmacology. 43 (1): 35–8. doi:10.1007/bf02280751. PMID 1505606.
- Nielsen-Kudsk JE, Buhl JS, Johannessen AC (February 1990). "Verapamil-induced inhibition of theophylline elimination in healthy humans". Pharmacology & Toxicology. 66 (2): 101–3. doi:10.1111/j.1600-0773.1990.tb00713.x. PMID 2315261.
- Gin AS, Stringer KA, Welage LS, Wilton JH, Matthews GE (August 1989). "The effect of verapamil on the pharmacokinetic disposition of theophylline in cigarette smokers". Journal of Clinical Pharmacology. 29 (8): 728–32. doi:10.1002/j.1552-4604.1989.tb03407.x. PMID 2778093.
- Sirmans SM, Pieper JA, Lalonde RL, Smith DG, Self TH (July 1988). "Effect of calcium channel blockers on theophylline disposition". Clinical Pharmacology and Therapeutics. 44 (1): 29–34. doi:10.1038/clpt.1988.108. PMID 3391002.
- Robson RA, Miners JO, Birkett DJ (March 1988). "Selective inhibitory effects of nifedipine and verapamil on oxidative metabolism: effects on theophylline". British Journal of Clinical Pharmacology. 25 (3): 397–400. doi:10.1111/j.1365-2125.1988.tb03319.x. PMC 1386365. PMID 3358901.
- Abernethy DR, Egan JM, Dickinson TH, Carrum G (March 1988). "Substrate-selective inhibition by verapamil and diltiazem: differential disposition of antipyrine and theophylline in humans". The Journal of Pharmacology and Experimental Therapeutics. 244 (3): 994–9. PMID 3252045.
- Katoh M, Nakajima M, Yamazaki H, Yokoi T (October 2000). "Inhibitory potencies of 1,4-dihydropyridine calcium antagonists to P-glycoprotein-mediated transport: comparison with the effects on CYP3A4". Pharmaceutical Research. 17 (10): 1189–97. doi:10.1023/a:1007568811691. PMID 11145223.
- Druglib.com
- Cockshott ID (2004). "Bicalutamide: clinical pharmacokinetics and metabolism". Clinical Pharmacokinetics. 43 (13): 855–78. doi:10.2165/00003088-200443130-00003. PMID 15509184.
- Matsumoto S, Yamazoe Y (February 2001). "Involvement of multiple human cytochromes P450 in the liver microsomal metabolism of astemizole and a comparison with terfenadine". British Journal of Clinical Pharmacology. 51 (2): 133–42. doi:10.1046/j.1365-2125.2001.01292.x. PMC 2014443. PMID 11259984.
- Ledger, Thomas; Tong, Winnie (1 August 2019). "Medicinal Mishap: Iatrogenic Cushing's syndrome with inhaled fluticason". Australian Prescriber. 42 (4): 139–140. doi:10.18773/austprescr.2019.040. PMC 6698236. PMID 31427846.
- Enzyme 1.14.13.32 at KEGG
- "Showing Protein Cytochrome P450 3A4 (HMDBP01018)". Human Metabolome Database. Retrieved 2017-08-05.
- Daly AK, King BP (May 2003). "Pharmacogenetics of oral anticoagulants". Pharmacogenetics. 13 (5): 247–52. doi:10.1097/00008571-200305000-00002. PMID 12724615.
- Lau WC, Waskell LA, Watkins PB, Neer CJ, Horowitz K, Hopp AS, Tait AR, Carville DG, Guyer KE, Bates ER (January 2003). "Atorvastatin reduces the ability of clopidogrel to inhibit platelet aggregation: a new drug-drug interaction". Circulation. 107 (1): 32–7. doi:10.1161/01.CIR.0000047060.60595.CC. PMID 12515739.
- Meyer MR, Bach M, Welter J, Bovens M, Turcant A, Maurer HH (July 2013). "Ketamine-derived designer drug methoxetamine: metabolism including isoenzyme kinetics and toxicological detectability using GC-MS and LC-(HR-)MSn". Analytical and Bioanalytical Chemistry. 405 (19): 6307–21. doi:10.1007/s00216-013-7051-6. PMID 23774830. S2CID 27966043.
- "LOSARTAN- losartan potassium tablet, film coated". DailyMed. 2018-12-26. Retrieved 2019-02-06.
- "Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers".
- Flower R, Rang HP, Dale MM, Ritter JM (2007). Rang & Dale's pharmacology. Edinburgh: Churchill Livingstone. ISBN 978-0-443-06911-6.
- Park JY, Kim KA, Kim SL (November 2003). "Chloramphenicol is a potent inhibitor of cytochrome P450 isoforms CYP2C19 and CYP3A4 in human liver microsomes". Antimicrobial Agents and Chemotherapy. 47 (11): 3464–9. doi:10.1128/AAC.47.11.3464-3469.2003. PMC 253795. PMID 14576103.
- Research, Center for Drug Evaluation and. "Drug Interactions & Labeling - Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers". www.fda.gov. Retrieved 2018-08-06.
- Karakurt, S. (2016). "Modulatory Effects of Rutin on the Expression of Cytochrome P450s and Antioxidant Enzymes in Human Hepatoma Cells". Acta Pharmaceutica (Zagreb, Croatia). 66 (4): 491–502. doi:10.1515/acph-2016-0046. PMID 27749250. S2CID 20274417.
- Ashour, M. L.; Youssef, F. S.; Gad, H. A.; Wink, M. (2017). "Inhibition of Cytochrome P450 (CYP3A4) Activity by Extracts From 57 Plants Used in Traditional Chinese Medicine (TCM)". Pharmacognosy Magazine. 13 (50): 300–308. doi:10.4103/0973-1296.204561. PMC 5421430. PMID 28539725.
- Product Information: ORAVIG(R) buccal tablets, miconazole buccal tablets. Praelia Pharmaceuticals, Inc (per FDA), Cary, NC, 2013.
- http://www.rxlist.com/valerian-page3/supplements.htm#Interactions%5B%5D
- Zhang W, Ramamoorthy Y, Tyndale RF, Sellers EM (June 2003). "Interaction of buprenorphine and its metabolite norbuprenorphine with cytochromes p450 in vitro". Drug Metabolism and Disposition. 31 (6): 768–72. doi:10.1124/dmd.31.6.768. PMID 12756210. S2CID 16229370.
- http://www.aapsj.org/abstracts/AM_2009/AAPS2009-001235.PDF%5B%5D Archived July 21, 2011, at the Wayback Machine
- Wen X, Wang JS, Kivistö KT, Neuvonen PJ, Backman JT (2001). "In vitro evaluation of valproic acid as an inhibitor of human cytochrome P450 isoforms: preferential inhibition of cytochrome P450 2C9 (CYP2C9)". Br J Clin Pharmacol. 52 (5): 547–53. doi:10.1046/j.0306-5251.2001.01474.x. PMC 2014611. PMID 11736863.
- Potent inhibition of human cytochrome P450 3A isoforms by cannabidiol: Role of phenolic hydroxyl groups in the resorcinol moiety
- Non-nucleoside reverse-transcriptase inhibitors have been shown to both induce and inhibit CYP3A4.
- Hidaka M, Fujita K, Ogikubo T, Yamasaki K, Iwakiri T, Okumura M, Kodama H, Arimori K (June 2004). "Potent inhibition by star fruit of human cytochrome P450 3A (CYP3A) activity". Drug Metabolism and Disposition. 32 (6): 581–3. doi:10.1124/dmd.32.6.581. PMID 15155547. S2CID 17392051.
- HCVadvocate.org Archived 2010-03-05 at the Wayback Machine
- Gaudineau C, Auclair K (May 2004). "Inhibition of human P450 enzymes by nicotinic acid and nicotinamide". Biochemical and Biophysical Research Communications. 317 (3): 950–6. doi:10.1016/j.bbrc.2004.03.137. PMID 15081432.
- Kimura Y, Ito H, Ohnishi R, Hatano T (January 2010). "Inhibitory effects of polyphenols on human cytochrome P450 3A4 and 2C9 activity". Food and Chemical Toxicology. 48 (1): 429–35. doi:10.1016/j.fct.2009.10.041. PMID 19883715.
Ginko Biloba has been shown to contain the potent inhibitor amentoflavone
- Lim, Y. P.; Ma, C. Y.; Liu, C. L.; Lin, Y. H.; Hu, M. L.; Chen, J. J.; Hung, D. Z.; Hsieh, W. T.; Huang, J. D. (2012). "Sesamin: A Naturally Occurring Lignan Inhibits CYP3A4 by Antagonizing the Pregnane X Receptor Activation". Evidence-Based Complementary and Alternative Medicine : Ecam. 2012: 242810. doi:10.1155/2012/242810. PMC 3356939. PMID 22645625.
- Bhardwaj RK, Glaeser H, Becquemont L, Klotz U, Gupta SK, Fromm MF (August 2002). "Piperine, a major constituent of black pepper, inhibits human P-glycoprotein and CYP3A4". The Journal of Pharmacology and Experimental Therapeutics. 302 (2): 645–50. doi:10.1124/jpet.102.034728. PMID 12130727. S2CID 7398172.
- Wen X, Wang JS, Neuvonen PJ, Backman JT (January 2002). "Isoniazid is a mechanism-based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes". European Journal of Clinical Pharmacology. 57 (11): 799–804. doi:10.1007/s00228-001-0396-3. PMID 11868802. S2CID 19299097.
- Savai J, Varghese A, Pandita N, Chintamaneni M (May 2015). "Investigation of CYP3A4 and CYP2D6 Interactions of Withania somnifera and Centella asiatica in Human Liver Microsomes". Phytotherapy Research. 29 (5): 785–90. doi:10.1002/ptr.5308. PMID 25684704.
- https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/203415lbl.pdf
- Johannessen SI, Landmark CJ (September 2010). "Antiepileptic drug interactions - principles and clinical implications". Current Neuropharmacology. 8 (3): 254–67. doi:10.2174/157015910792246254. PMC 3001218. PMID 21358975.
- Nallani SC, Glauser TA, Hariparsad N, Setchell K, Buckley DJ, Buckley AR, Desai PB (December 2003). "Dose-dependent induction of cytochrome P450 (CYP) 3A4 and activation of pregnane X receptor by topiramate". Epilepsia. 44 (12): 1521–8. doi:10.1111/j.0013-9580.2003.06203.x. PMID 14636322.
- Chae YJ, Cho KH, Yoon IS, Noh CK, Lee HJ, Park Y, Ji E, Seo MD, Maeng HJ (January 2016). "Vitamin D Receptor-Mediated Upregulation of CYP3A4 and MDR1 by Quercetin in Caco-2 cells". Planta Medica. 82 (1–2): 121–30. doi:10.1055/s-0035-1557898. PMID 26366751.
- Han EH, Kim HG, Choi JH, Jang YJ, Lee SS, Kwon KI, Kim E, Noh K, Jeong TC, Hwang YP, Chung YC, Kang W, Jeong HG (May 2012). "Capsaicin induces CYP3A4 expression via pregnane X receptor and CCAAT/enhancer-binding protein β activation". Molecular Nutrition & Food Research. 56 (5): 797–809. doi:10.1002/mnfr.201100697. PMID 22648626. S2CID 26584141.
External links
- PharmGKB: Annotated PGx Gene Information for CYP3A4
- CYP3A4 substrate prediction
- Human CYP3A4 genome location and CYP3A4 gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P08684 (Cytochrome P450 3A4) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.