Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. The Hsp70s are an important part of the cell's machinery for protein folding, and help to protect cells from stress.[2][3]
| Hsp70 protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
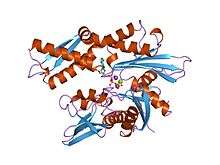 Structure of the ATPase fragment of a 70K heat-shock cognate protein.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | HSP70 | ||||||||
| Pfam | PF00012 | ||||||||
| Pfam clan | CL0108 | ||||||||
| InterPro | IPR013126 | ||||||||
| PROSITE | PDOC00269 | ||||||||
| SCOPe | 3hsc / SUPFAM | ||||||||
| |||||||||
Discovery
Members of the Hsp70 family are very strongly upregulated by heat stress and toxic chemicals, particularly heavy metals such as arsenic, cadmium, copper, mercury, etc. Hsp70 was originally discovered by FM Ritossa in the 1960s when a lab worker accidentally boosted the incubation temperature of Drosophila (fruit flies). When examining the chromosomes, Ritossa found a "puffing pattern" that indicated the elevated gene transcription of an unknown protein.[4][5] This was later described as the "Heat Shock Response" and the proteins were termed the "Heat Shock Proteins" (Hsps).
Structure

The Hsp70 proteins have three major functional domains:
- N-terminal ATPase domain – binds ATP (Adenosine triphosphate) and hydrolyzes it to ADP (Adenosine diphosphate). The NBD consists of two lobes with a deep cleft between them, at the bottom of which nucleotide (ATP and ADP) binds. The exchange of ATP and ADP leads to conformational changes in the other two domains.
- Substrate binding domain – is composed of a 15 kDa β sheet subdomain and a 10 kDa helical subdomain. The β sheet subdomain consists of stranded β sheets with upward protruding loops, as a typical β barrel, which enclose the peptide backbone of the substrate. SBD contains a groove with an affinity for neutral, hydrophobic amino acid residues. The groove is long enough to interact with peptides up to seven residues in length.
- C-terminal domain – rich in alpha helical structure acts as a 'lid' for the substrate binding domain. The helical subdomain consists of five helices, with two helices packed against two sides of the β sheet subdomain, stabilizing the inner structure. In addition, one of the helix forms a salt bridge and several hydrogen bonds to the outer Loops, thereby closing the substrate-binding pocket like a lid. Three helices in this domain form another hydrophobic core which may be stabilization of the "lid". When an Hsp70 protein is ATP bound, the lid is open and peptides bind and release relatively rapidly. When Hsp70 proteins are ADP bound, the lid is closed, and peptides are tightly bound to the substrate binding domain.[6]
Function and regulation
The Hsp70 system interacts with extended peptide segments of proteins as well as partially folded proteins to cause aggregation of proteins in key pathways to deregulate activity.[7] When not interacting with a substrate peptide, Hsp70 is usually in an ATP bound state. Hsp70 by itself is characterized by a very weak ATPase activity, such that spontaneous hydrolysis will not occur for many minutes. As newly synthesized proteins emerge from the ribosomes, the substrate binding domain of Hsp70 recognizes sequences of hydrophobic amino acid residues, and interacts with them. This spontaneous interaction is reversible, and in the ATP bound state Hsp70 may relatively freely bind and release peptides. However, the presence of a peptide in the binding domain stimulates the ATPase activity of Hsp70, increasing its normally slow rate of ATP hydrolysis. When ATP is hydrolyzed to ADP the binding pocket of Hsp70 closes, tightly binding the now-trapped peptide chain. Further speeding ATP hydrolysis are the so-called J-domain cochaperones: primarily Hsp40 in eukaryotes, and DnaJ in prokaryotes. These cochaperones dramatically increase the ATPase activity of Hsp70 in the presence of interacting peptides.
By binding tightly to partially synthesized peptide sequences (incomplete proteins), Hsp70 prevents them from aggregating and being rendered nonfunctional. Once the entire protein is synthesized, a nucleotide exchange factor (prokaryotic GrpE, eukaryotic BAG1 and HspBP1 are among those which have been identified) stimulates the release of ADP and binding of fresh ATP, opening the binding pocket. The protein is then free to fold on its own, or to be transferred to other chaperones for further processing.[8] HOP (the Hsp70/Hsp90 Organizing Protein) can bind to both Hsp70 and Hsp90 at the same time, and mediates the transfer of peptides from Hsp70 to Hsp90.[9]
Hsp70 also aids in transmembrane transport of proteins, by stabilizing them in a partially folded state. It is also known to be phosphorylated[10] which regulates several of its functions.[11][12][13]
Hsp70 proteins can act to protect cells from thermal or oxidative stress. These stresses normally act to damage proteins, causing partial unfolding and possible aggregation. By temporarily binding to hydrophobic residues exposed by stress, Hsp70 prevents these partially denatured proteins from aggregating, and inhibits them from refolding. Low ATP is characteristic of heat shock and sustained binding is seen as aggregation suppression, while recovery from heat shock involves substrate binding and nucleotide cycling. In a thermophile anaerobe (Thermotoga maritima) the Hsp70 demonstrates redox sensitive binding to model peptides, suggesting a second mode of binding regulation based on oxidative stress.
Hsp70 seems to be able to participate in disposal of damaged or defective proteins. Interaction with CHIP (Carboxyl-terminus of Hsp70 Interacting Protein)–an E3 ubiquitin ligase–allows Hsp70 to pass proteins to the cell's ubiquitination and proteolysis pathways.[14]
Finally, in addition to improving overall protein integrity, Hsp70 directly inhibits apoptosis.[15] One hallmark of apoptosis is the release of cytochrome c, which then recruits Apaf-1 and dATP/ATP into an apoptosome complex. This complex then cleaves procaspase-9, activating caspase-9 and eventually inducing apoptosis via caspase-3 activation. Hsp70 inhibits this process by blocking the recruitment of procaspase-9 to the Apaf-1/dATP/cytochrome c apoptosome complex. It does not bind directly to the procaspase-9 binding site, but likely induces a conformational change that renders procaspase-9 binding less favorable. Hsp70 is shown to interact with Endoplasmic reticulum stress sensor protein IRE1alpha thereby protecting the cells from ER stress - induced apoptosis. This interaction prolonged the splicing of XBP-1 mRNA thereby inducing transcriptional upregulation of targets of spliced XBP-1 like EDEM1, ERdj4 and P58IPK rescuing the cells from apoptosis.[16] Other studies suggest that Hsp70 may play an anti-apoptotic role at other steps, but is not involved in Fas-ligand-mediated apoptosis (although Hsp 27 is). Therefore, Hsp70 not only saves important components of the cell (the proteins) but also directly saves the cell as a whole. Considering that stress-response proteins (like Hsp70) evolved before apoptotic machinery, Hsp70's direct role in inhibiting apoptosis provides an interesting evolutionary picture of how more recent (apoptotic) machinery accommodated previous machinery (Hsps), thus aligning the improved integrity of a cell's proteins with the improved chances of that particular cell's survival.
Cancer
Hsp70 is overexpressed in malignant melanoma[17] and underexpressed in renal cell cancer.[18][19]
Expression in skin tissue
Both HSP70 and HSP47 were shown to be expressed in dermis and epidermis following laser irradiation, and the spatial and temporal changes in HSP expression patterns define the laser-induced thermal damage zone and the process of healing in tissues. HSP70 may define biochemically the thermal damage zone in which cells are targeted for destruction, and HSP47 may illustrate the process of recovery from thermally induced damage.[20]
Family members
Prokaryotes express three Hsp70 proteins: DnaK, HscA (Hsc66), and HscC (Hsc62).[21]
Eukaryotic organisms express several slightly different Hsp70 proteins. All share the common domain structure, but each has a unique pattern of expression or subcellular localization. These are, among others:
- Hsc70 (Hsp73/HSPA8) is a constitutively expressed chaperone protein. It typically makes up one to three percent of total cellular protein.
- Hsp70 (encoded by three very closely related paralogs: HSPA1A, HSPA1B, and HSPA1L) is a stress-induced protein. High levels can be produced by cells in response to hyperthermia, oxidative stress, and changes in pH.
- Binding immunoglobulin protein (BiP or Grp78) is a protein localized to the endoplasmic reticulum. It is involved in protein folding there, and can be upregulated in response to stress or starvation.
- mtHsp70 or Grp75 is the mitochondrial Hsp70.
The following is a list of human Hsp70 genes and their corresponding proteins:[2]
| gene | protein | synonyms | subcellular location |
|---|---|---|---|
| HSPA1A | Hsp70 | HSP70-1, Hsp72 | Nuc/Cyto |
| HSPA1B | Hsp70 | HSP70-2 | Nuc/Cyto |
| HSPA1L | Hsp70 | ? | |
| HSPA2 | Hsp70-2 | ? | |
| HSPA5 | Hsp70-5 | BiP/Grp78 | ER |
| HSPA6 | Hsp70-6 | ? | |
| HSPA7 | Hsp70-7 | ? | |
| HSPA8 | Hsp70-8 | Hsc70 | Nuc/Cyto |
| HSPA9 | Hsp70-9 | Grp75/mtHsp70 | Mito |
| HSPA12A | Hsp70-12a | ? | |
| HSPA14 | Hsp70-14 | ? |
Hsp110
The Hsp70 superfamily also includes a family of Hsp110/Grp170 (Sse) proteins, which are larger proteins related to Hsp70.[22] The Hsp110 family of proteins have divergent functions: yeast Sse1p has little ATPase activity but is a chaperone on its own as well as a nucleotide exchange factor for Hsp70, while the closely related Sse2p has little unfoldase activity.[8]
The following is a list of currently named human HSP110 genes. HSPH2-4 are proposed names and the current name is linked:[22]
| gene | synonyms | subcellular location |
|---|---|---|
| HSPH1 | HSP105 | Cyto |
| HSPH2 | HSPA4; APG-2; HSP110 | Cyto |
| HSPH3 | HSPA4L; APG-1 | Nuc |
| HSPH4 | HYOU1/Grp170; ORP150; HSP12A | ER |
See also
- Mitochondrial import stimulation factor
- Heat shock protein 70 (Hsp70) internal ribosome entry site (IRES)
References
- Flaherty KM, DeLuca-Flaherty C, McKay DB (August 1990). "Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein". Nature. 346 (6285): 623–8. Bibcode:1990Natur.346..623F. doi:10.1038/346623a0. PMID 2143562.
- Tavaria M, Gabriele T, Kola I, Anderson RL (April 1996). "A hitchhiker's guide to the human Hsp70 family". Cell Stress & Chaperones. 1 (1): 23–8. doi:10.1379/1466-1268(1996)001<0023:AHSGTT>2.3.CO;2. PMC 313013. PMID 9222585.
- Morano KA (October 2007). "New tricks for an old dog: the evolving world of Hsp70". Annals of the New York Academy of Sciences. 1113 (1): 1–14. Bibcode:2007NYASA1113....1M. doi:10.1196/annals.1391.018. PMID 17513460.
- Ritossa F (1962). "A new puffing pattern induced by temperature shock and DNP in drosophila". Cellular and Molecular Life Sciences. 18 (12): 571–573. doi:10.1007/BF02172188.
- Ritossa F (June 1996). "Discovery of the heat shock response". Cell Stress & Chaperones. 1 (2): 97–8. doi:10.1379/1466-1268(1996)001<0097:DOTHSR>2.3.CO;2. PMC 248460. PMID 9222594.
- Mayer MP (August 2010). "Gymnastics of molecular chaperones". Molecular Cell. 39 (3): 321–31. doi:10.1016/j.molcel.2010.07.012. PMID 20705236.
- Mashaghi A, Bezrukavnikov S, Minde DP, Wentink AS, Kityk R, Zachmann-Brand B, Mayer MP, Kramer G, Bukau B, Tans SJ (November 2016). "Alternative modes of client binding enable functional plasticity of Hsp70". Nature. 539 (7629): 448–451. Bibcode:2016Natur.539..448M. doi:10.1038/nature20137. PMID 27783598.
- Bracher A, Verghese J (2015). "GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones". Sub-Cellular Biochemistry. Subcellular Biochemistry. 78: 1–33. doi:10.1007/978-3-319-11731-7_1. ISBN 978-3-319-11730-0. PMID 25487014.
- Wegele H, Müller L, Buchner J (2004). Hsp70 and Hsp90 – a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. Reviews of Physiology, Biochemistry and Pharmacology. 151. pp. 1–44. doi:10.1007/s10254-003-0021-1. ISBN 978-3-540-22096-1. PMID 14740253.
- Cvoro A, Dundjerski J, Trajković D, Matić G (1999-04-01). "The level and phosphorylation of Hsp70 in the rat liver cytosol after adrenalectomy and hyperthermia". Cell Biology International. 23 (4): 313–20. doi:10.1006/cbir.1998.0247. PMID 10600240.
- Gao T, Newton AC (August 2002). "The turn motif is a phosphorylation switch that regulates the binding of Hsp70 to protein kinase C". The Journal of Biological Chemistry. 277 (35): 31585–92. doi:10.1074/jbc.M204335200. PMID 12080070.
- Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, Kron SJ (December 2012). "CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression". Cell. 151 (6): 1308–18. doi:10.1016/j.cell.2012.10.051. PMC 3778871. PMID 23217712.
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B (June 2013). "C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances". Oncogene. 32 (25): 3101–10. doi:10.1038/onc.2012.314. PMID 22824801.
- Lüders J, Demand J, Höhfeld J (February 2000). "The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome". The Journal of Biological Chemistry. 275 (7): 4613–7. doi:10.1074/jbc.275.7.4613. PMID 10671488.
- Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR (August 2000). "Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome". Nature Cell Biology. 2 (8): 469–75. doi:10.1038/35019501. PMID 10934466.
- Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A (July 2010). Kelly JW (ed.). "HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction". PLoS Biology. 8 (7): e1000410. doi:10.1371/journal.pbio.1000410. PMC 2897763. PMID 20625543.
- Ricaniadis N, Kataki A, Agnantis N, Androulakis G, Karakousis CP (February 2001). "Long-term prognostic significance of HSP-70, c-myc and HLA-DR expression in patients with malignant melanoma". European Journal of Surgical Oncology. 27 (1): 88–93. doi:10.1053/ejso.1999.1018. PMID 11237497.
- Ramp U, Mahotka C, Heikaus S, Shibata T, Grimm MO, Willers R, Gabbert HE (October 2007). "Expression of heat shock protein 70 in renal cell carcinoma and its relation to tumor progression and prognosis". Histology and Histopathology. 22 (10): 1099–107. doi:10.14670/HH-22.1099. PMID 17616937.
- Sherman M, Multhoff G (October 2007). "Heat shock proteins in cancer". Annals of the New York Academy of Sciences. 1113 (1): 192–201. Bibcode:2007NYASA1113..192S. doi:10.1196/annals.1391.030. PMID 17978282.
- Sajjadi AY, Mitra K, Grace M (October 2013). "Expression of heat shock proteins 70 and 47 in tissues following short-pulse laser irradiation: assessment of thermal damage and healing" (PDF). Medical Engineering & Physics. 35 (10): 1406–14. doi:10.1016/j.medengphy.2013.03.011. PMID 23587755.
- Yoshimune K, Yoshimura T, Nakayama T, Nishino T, Esaki N (May 2002). "Hsc62, Hsc56, and GrpE, the third Hsp70 chaperone system of Escherichia coli". Biochemical and Biophysical Research Communications. 293 (5): 1389–95. doi:10.1016/S0006-291X(02)00403-5. PMID 12054669.
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE (January 2009). "Guidelines for the nomenclature of the human heat shock proteins". Cell Stress & Chaperones. 14 (1): 105–11. doi:10.1007/s12192-008-0068-7. PMC 2673902. PMID 18663603.
External links
- HSP70+Heat-Shock+Proteins at the US National Library of Medicine Medical Subject Headings (MeSH)