Microevolution
Microevolution is the change in allele frequencies that occurs over time within a population.[1] This change is due to four different processes: mutation, selection (natural and artificial), gene flow and genetic drift. This change happens over a relatively short (in evolutionary terms) amount of time compared to the changes termed macroevolution.
| Part of a series on |
| Evolutionary biology |
|---|
 |
|
Key topics |
|
Fields and applications
|
|
Social implications
|
|
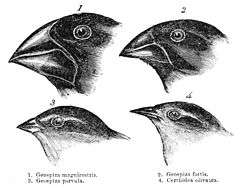
Population genetics is the branch of biology that provides the mathematical structure for the study of the process of microevolution. Ecological genetics concerns itself with observing microevolution in the wild. Typically, observable instances of evolution are examples of microevolution; for example, bacterial strains that have antibiotic resistance.
Microevolution may lead to speciation, which provides the raw material for macroevolution.[2][3]
Difference from macroevolution
Macroevolution is guided by sorting of interspecific variation ("species selection"[2]), as opposed to sorting of intraspecific variation in microevolution.[3] Species selection may occur as (a) effect-macroevolution, where organism-level traits (aggregate traits) affect speciation and extinction rates, and (b) strict-sense species selection, where species-level traits (e.g. geographical range) affect speciation and extinction rates.[4] Macroevolution does not produce evolutionary novelties, but it determines their proliferation within the clades in which they evolved, and it adds species-level traits as non-organismic factors of sorting to this process.[3]
Four processes
Mutation

Mutations are changes in the DNA sequence of a cell's genome and are caused by radiation, viruses, transposons and mutagenic chemicals, as well as errors that occur during meiosis or DNA replication.[5][6][7] Errors are introduced particularly often in the process of DNA replication, in the polymerization of the second strand. These errors can also be induced by the organism itself, by cellular processes such as hypermutation. Mutations can affect the phenotype of an organism, especially if they occur within the protein coding sequence of a gene. Error rates are usually very low—1 error in every 10–100 million bases—due to the proofreading ability of DNA polymerases.[8][9] (Without proofreading error rates are a thousandfold higher; because many viruses rely on DNA and RNA polymerases that lack proofreading ability, they experience higher mutation rates.) Processes that increase the rate of changes in DNA are called mutagenic: mutagenic chemicals promote errors in DNA replication, often by interfering with the structure of base-pairing, while UV radiation induces mutations by causing damage to the DNA structure.[10] Chemical damage to DNA occurs naturally as well, and cells use DNA repair mechanisms to repair mismatches and breaks in DNA—nevertheless, the repair sometimes fails to return the DNA to its original sequence.
In organisms that use chromosomal crossover to exchange DNA and recombine genes, errors in alignment during meiosis can also cause mutations.[11] Errors in crossover are especially likely when similar sequences cause partner chromosomes to adopt a mistaken alignment making some regions in genomes more prone to mutating in this way. These errors create large structural changes in DNA sequence—duplications, inversions or deletions of entire regions, or the accidental exchanging of whole parts between different chromosomes (called translocation).
Mutation can result in several different types of change in DNA sequences; these can either have no effect, alter the product of a gene, or prevent the gene from functioning. Studies in the fly Drosophila melanogaster suggest that if a mutation changes a protein produced by a gene, this will probably be harmful, with about 70 percent of these mutations having damaging effects, and the remainder being either neutral or weakly beneficial.[12] Due to the damaging effects that mutations can have on cells, organisms have evolved mechanisms such as DNA repair to remove mutations.[5] Therefore, the optimal mutation rate for a species is a trade-off between costs of a high mutation rate, such as deleterious mutations, and the metabolic costs of maintaining systems to reduce the mutation rate, such as DNA repair enzymes.[13] Viruses that use RNA as their genetic material have rapid mutation rates,[14] which can be an advantage since these viruses will evolve constantly and rapidly, and thus evade the defensive responses of e.g. the human immune system.[15]
Mutations can involve large sections of DNA becoming duplicated, usually through genetic recombination.[16] These duplications are a major source of raw material for evolving new genes, with tens to hundreds of genes duplicated in animal genomes every million years.[17] Most genes belong to larger families of genes of shared ancestry.[18] Novel genes are produced by several methods, commonly through the duplication and mutation of an ancestral gene, or by recombining parts of different genes to form new combinations with new functions.[19][20]
Here, domains act as modules, each with a particular and independent function, that can be mixed together to produce genes encoding new proteins with novel properties.[21] For example, the human eye uses four genes to make structures that sense light: three for color vision and one for night vision; all four arose from a single ancestral gene.[22] Another advantage of duplicating a gene (or even an entire genome) is that this increases redundancy; this allows one gene in the pair to acquire a new function while the other copy performs the original function.[23][24] Other types of mutation occasionally create new genes from previously noncoding DNA.[25][26]
Selection
Selection is the process by which heritable traits that make it more likely for an organism to survive and successfully reproduce become more common in a population over successive generations.
It is sometimes valuable to distinguish between naturally occurring selection, natural selection, and selection that is a manifestation of choices made by humans, artificial selection. This distinction is rather diffuse. Natural selection is nevertheless the dominant part of selection.
The natural genetic variation within a population of organisms means that some individuals will survive more successfully than others in their current environment. Factors which affect reproductive success are also important, an issue which Charles Darwin developed in his ideas on sexual selection.
Natural selection acts on the phenotype, or the observable characteristics of an organism, but the genetic (heritable) basis of any phenotype which gives a reproductive advantage will become more common in a population (see allele frequency). Over time, this process can result in adaptations that specialize organisms for particular ecological niches and may eventually result in the speciation (the emergence of new species).
Natural selection is one of the cornerstones of modern biology. The term was introduced by Darwin in his groundbreaking 1859 book On the Origin of Species,[27] in which natural selection was described by analogy to artificial selection, a process by which animals and plants with traits considered desirable by human breeders are systematically favored for reproduction. The concept of natural selection was originally developed in the absence of a valid theory of heredity; at the time of Darwin's writing, nothing was known of modern genetics. The union of traditional Darwinian evolution with subsequent discoveries in classical and molecular genetics is termed the modern evolutionary synthesis. Natural selection remains the primary explanation for adaptive evolution.
Genetic drift
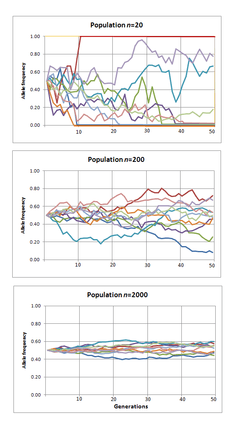
Genetic drift is the change in the relative frequency in which a gene variant (allele) occurs in a population due to random sampling. That is, the alleles in the offspring in the population are a random sample of those in the parents. And chance has a role in determining whether a given individual survives and reproduces. A population's allele frequency is the fraction or percentage of its gene copies compared to the total number of gene alleles that share a particular form.[28]
Genetic drift is an evolutionary process which leads to changes in allele frequencies over time. It may cause gene variants to disappear completely, and thereby reduce genetic variability. In contrast to natural selection, which makes gene variants more common or less common depending on their reproductive success,[29] the changes due to genetic drift are not driven by environmental or adaptive pressures, and may be beneficial, neutral, or detrimental to reproductive success.
The effect of genetic drift is larger in small populations, and smaller in large populations. Vigorous debates wage among scientists over the relative importance of genetic drift compared with natural selection. Ronald Fisher held the view that genetic drift plays at the most a minor role in evolution, and this remained the dominant view for several decades. In 1968 Motoo Kimura rekindled the debate with his neutral theory of molecular evolution which claims that most of the changes in the genetic material are caused by genetic drift.[30] The predictions of neutral theory, based on genetic drift, do not fit recent data on whole genomes well: these data suggest that the frequencies of neutral alleles change primarily due to selection at linked sites, rather than due to genetic drift by means of sampling error.[31]
Gene flow
Gene flow is the exchange of genes between populations, which are usually of the same species.[32] Examples of gene flow within a species include the migration and then breeding of organisms, or the exchange of pollen. Gene transfer between species includes the formation of hybrid organisms and horizontal gene transfer.
Migration into or out of a population can change allele frequencies, as well as introducing genetic variation into a population. Immigration may add new genetic material to the established gene pool of a population. Conversely, emigration may remove genetic material. As barriers to reproduction between two diverging populations are required for the populations to become new species, gene flow may slow this process by spreading genetic differences between the populations. Gene flow is hindered by mountain ranges, oceans and deserts or even man-made structures such as the Great Wall of China, which has hindered the flow of plant genes.[33]
Depending on how far two species have diverged since their most recent common ancestor, it may still be possible for them to produce offspring, as with horses and donkeys mating to produce mules.[34] Such hybrids are generally infertile, due to the two different sets of chromosomes being unable to pair up during meiosis. In this case, closely related species may regularly interbreed, but hybrids will be selected against and the species will remain distinct. However, viable hybrids are occasionally formed and these new species can either have properties intermediate between their parent species, or possess a totally new phenotype.[35] The importance of hybridization in creating new species of animals is unclear, although cases have been seen in many types of animals,[36] with the gray tree frog being a particularly well-studied example.[37]
Hybridization is, however, an important means of speciation in plants, since polyploidy (having more than two copies of each chromosome) is tolerated in plants more readily than in animals.[38][39] Polyploidy is important in hybrids as it allows reproduction, with the two different sets of chromosomes each being able to pair with an identical partner during meiosis.[40] Polyploid hybrids also have more genetic diversity, which allows them to avoid inbreeding depression in small populations.[41]
Horizontal gene transfer is the transfer of genetic material from one organism to another organism that is not its offspring; this is most common among bacteria.[42] In medicine, this contributes to the spread of antibiotic resistance, as when one bacteria acquires resistance genes it can rapidly transfer them to other species.[43] Horizontal transfer of genes from bacteria to eukaryotes such as the yeast Saccharomyces cerevisiae and the adzuki bean beetle Callosobruchus chinensis may also have occurred.[44][45] An example of larger-scale transfers are the eukaryotic bdelloid rotifers, which appear to have received a range of genes from bacteria, fungi, and plants.[46] Viruses can also carry DNA between organisms, allowing transfer of genes even across biological domains.[47] Large-scale gene transfer has also occurred between the ancestors of eukaryotic cells and prokaryotes, during the acquisition of chloroplasts and mitochondria.[48]
Gene flow is the transfer of alleles from one population to another.
Migration into or out of a population may be responsible for a marked change in allele frequencies. Immigration may also result in the addition of new genetic variants to the established gene pool of a particular species or population.
There are a number of factors that affect the rate of gene flow between different populations. One of the most significant factors is mobility, as greater mobility of an individual tends to give it greater migratory potential. Animals tend to be more mobile than plants, although pollen and seeds may be carried great distances by animals or wind.
Maintained gene flow between two populations can also lead to a combination of the two gene pools, reducing the genetic variation between the two groups. It is for this reason that gene flow strongly acts against speciation, by recombining the gene pools of the groups, and thus, repairing the developing differences in genetic variation that would have led to full speciation and creation of daughter species.
For example, if a species of grass grows on both sides of a highway, pollen is likely to be transported from one side to the other and vice versa. If this pollen is able to fertilise the plant where it ends up and produce viable offspring, then the alleles in the pollen have effectively been able to move from the population on one side of the highway to the other.
Origin and extended use of the term
Origin
The term microevolution was first used by botanist Robert Greenleaf Leavitt in the journal Botanical Gazette in 1909, addressing what he called the "mystery" of how formlessness gives rise to form.[49]
- ..The production of form from formlessness in the egg-derived individual, the multiplication of parts and the orderly creation of diversity among them, in an actual evolution, of which anyone may ascertain the facts, but of which no one has dissipated the mystery in any significant measure. This microevolution forms an integral part of the grand evolution problem and lies at the base of it, so that we shall have to understand the minor process before we can thoroughly comprehend the more general one...
However, Leavitt was using the term to describe what we would now call developmental biology; it was not until Russian Entomologist Yuri Filipchenko used the terms "macroevolution" and "microevolution" in 1927 in his German language work, Variabilität und Variation, that it attained its modern usage. The term was later brought into the English-speaking world by Filipchenko's student Theodosius Dobzhansky in his book Genetics and the Origin of Species (1937).[1]
Use in creationism
In young Earth creationism and baraminology a central tenet is that evolution can explain diversity in a limited number of created kinds which can interbreed (which they call "microevolution") while the formation of new "kinds" (which they call "macroevolution") is impossible.[50][51] This acceptance of "microevolution" only within a "kind" is also typical of old Earth creationism.[52]
Scientific organizations such as the American Association for the Advancement of Science describe microevolution as small scale change within species, and macroevolution as the formation of new species, but otherwise not being different from microevolution. In macroevolution, an accumulation of microevolutionary changes leads to speciation.[53] The main difference between the two processes is that one occurs within a few generations, whilst the other takes place over thousands of years (i.e. a quantitative difference).[54] Essentially they describe the same process; although evolution beyond the species level results in beginning and ending generations which could not interbreed, the intermediate generations could.
Opponents to creationism argue that changes in the number of chromosomes can be accounted for by intermediate stages in which a single chromosome divides in generational stages, or multiple chromosomes fuse, and cite the chromosome difference between humans and the other great apes as an example.[55] Creationists insist that since the actual divergence between the other great apes and humans was not observed, the evidence is circumstantial.
Describing the fundamental similarity between macro and microevolution in his authoritative textbook "Evolutionary Biology," biologist Douglas Futuyma writes,
One of the most important tenets of the theory forged during the Evolutionary Synthesis of the 1930s and 1940s was that "macroevolutionary" differences among organisms - those that distinguish higher taxa - arise from the accumulation of the same kinds of genetic differences that are found within species. Opponents of this point of view believed that "macroevolution" is qualitatively different from "microevolution" within species, and is based on a totally different kind of genetic and developmental patterning... Genetic studies of species differences have decisively disproved [this] claim. Differences between species in morphology, behavior, and the processes that underlie reproductive isolation all have the same genetic properties as variation within species: they occupy consistent chromosomal positions, they may be polygenic or based on few genes, they may display additive, dominant, or epistatic effects, and they can in some instances be traced to specifiable differences in proteins or DNA nucleotide sequences. The degree of reproductive isolation between populations, whether prezygotic or postzygotic, varies from little or none to complete. Thus, reproductive isolation, like the divergence of any other character, evolves in most cases by the gradual substitution of alleles in populations.
— Douglas Futuyma, "Evolutionary Biology" (1998), pp.477-8[56]
Contrary to the claims of some antievolution proponents, evolution of life forms beyond the species level (i.e. speciation) has indeed been observed and documented by scientists on numerous occasions.[57] In creation science, creationists accepted speciation as occurring within a "created kind" or "baramin", but objected to what they called "third level-macroevolution" of a new genus or higher rank in taxonomy. There is ambiguity in the ideas as to where to draw a line on "species", "created kinds", and what events and lineages fall within the rubric of microevolution or macroevolution.[58]
See also
- Punctuated equilibrium - due to gene flow, major evolutionary changes may be rare
References
- Microevolution: What is microevolution?
- Stanley, S. M. (1 February 1975). "A theory of evolution above the species level". Proceedings of the National Academy of Sciences. 72 (2): 646–650. Bibcode:1975PNAS...72..646S. doi:10.1073/pnas.72.2.646. ISSN 0027-8424. PMC 432371. PMID 1054846.
- Hautmann, Michael (2020). "What is macroevolution?". Palaeontology. 63 (1): 1–11. doi:10.1111/pala.12465. ISSN 0031-0239.
- Jablonski, David (December 2008). "Species Selection: Theory and Data". Annual Review of Ecology, Evolution, and Systematics. 39 (1): 501–524. doi:10.1146/annurev.ecolsys.39.110707.173510. ISSN 1543-592X.
- Bertram J (2000). "The molecular biology of cancer". Mol. Aspects Med. 21 (6): 167–223. doi:10.1016/S0098-2997(00)00007-8. PMID 11173079.
- Aminetzach YT, Macpherson JM, Petrov DA; MacPherson; Petrov (2005). "Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila". Science. 309 (5735): 764–7. Bibcode:2005Sci...309..764A. doi:10.1126/science.1112699. PMID 16051794.CS1 maint: multiple names: authors list (link)
- Burrus V, Waldor M; Waldor (2004). "Shaping bacterial genomes with integrative and conjugative elements". Res. Microbiol. 155 (5): 376–86. doi:10.1016/j.resmic.2004.01.012. PMID 15207870.
- Griffiths, Anthony J. F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M., eds. (2000). "Spontaneous mutations". An Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman. ISBN 978-0-7167-3520-5.
- Freisinger, E; Grollman, AP; Miller, H; Kisker, C (2004). "Lesion (in)tolerance reveals insights into DNA replication fidelity". The EMBO Journal. 23 (7): 1494–505. doi:10.1038/sj.emboj.7600158. PMC 391067. PMID 15057282.
- Griffiths, Anthony J. F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M., eds. (2000). "Induced mutations". An Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman. ISBN 978-0-7167-3520-5.
- Griffiths, Anthony J. F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M., eds. (2000). "Chromosome Mutation I: Changes in Chromosome Structure: Introduction". An Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman. ISBN 978-0-7167-3520-5.
- Sawyer SA, Parsch J, Zhang Z, Hartl DL; Parsch; Zhang; Hartl (2007). "Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila". Proc. Natl. Acad. Sci. U.S.A. 104 (16): 6504–10. Bibcode:2007PNAS..104.6504S. doi:10.1073/pnas.0701572104. PMC 1871816. PMID 17409186.CS1 maint: multiple names: authors list (link)
- Sniegowski P, Gerrish P, Johnson T, Shaver A; Gerrish; Johnson; Shaver (2000). "The evolution of mutation rates: separating causes from consequences". BioEssays. 22 (12): 1057–66. doi:10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. PMID 11084621.CS1 maint: multiple names: authors list (link)
- Drake JW, Holland JJ; Holland (1999). "Mutation rates among RNA viruses". Proc. Natl. Acad. Sci. U.S.A. 96 (24): 13910–3. Bibcode:1999PNAS...9613910D. doi:10.1073/pnas.96.24.13910. PMC 24164. PMID 10570172.
- Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S; Spindler; Horodyski; Grabau; Nichol; Vandepol (1982). "Rapid evolution of RNA genomes". Science. 215 (4540): 1577–85. Bibcode:1982Sci...215.1577H. doi:10.1126/science.7041255. PMID 7041255.CS1 maint: multiple names: authors list (link)
- Hastings, P J; Lupski, JR; Rosenberg, SM; Ira, G (2009). "Mechanisms of change in gene copy number". Nature Reviews Genetics. 10 (8): 551–564. doi:10.1038/nrg2593. PMC 2864001. PMID 19597530.
- Carroll SB, Grenier J, Weatherbee SD (2005). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Second Edition. Oxford: Blackwell Publishing. ISBN 978-1-4051-1950-4.
- Harrison P, Gerstein M; Gerstein (2002). "Studying genomes through the aeons: protein families, pseudogenes and proteome evolution". J Mol Biol. 318 (5): 1155–74. doi:10.1016/S0022-2836(02)00109-2. PMID 12083509.
- Orengo CA, Thornton JM; Thornton (2005). "Protein families and their evolution-a structural perspective". Annu. Rev. Biochem. 74: 867–900. doi:10.1146/annurev.biochem.74.082803.133029. PMID 15954844.
- Long M, Betrán E, Thornton K, Wang W; Betrán; Thornton; Wang (November 2003). "The origin of new genes: glimpses from the young and old". Nature Reviews Genetics. 4 (11): 865–75. doi:10.1038/nrg1204. PMID 14634634.CS1 maint: multiple names: authors list (link)
- Wang M, Caetano-Anollés G; Caetano-Anollés (2009). "The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world". Structure. 17 (1): 66–78. doi:10.1016/j.str.2008.11.008. PMID 19141283.
- Bowmaker JK (1998). "Evolution of colour vision in vertebrates". Eye (London, England). 12 (Pt 3b): 541–7. doi:10.1038/eye.1998.143. PMID 9775215.
- Gregory TR, Hebert PD; Hebert (1999). "The modulation of DNA content: proximate causes and ultimate consequences". Genome Res. 9 (4): 317–24. doi:10.1101/gr.9.4.317 (inactive 31 May 2020). PMID 10207154.
- Hurles M (July 2004). "Gene duplication: the genomic trade in spare parts". PLOS Biol. 2 (7): E206. doi:10.1371/journal.pbio.0020206. PMC 449868. PMID 15252449.
- Liu N, Okamura K, Tyler DM; Okamura; Tyler; Phillips; Chung; Lai (2008). "The evolution and functional diversification of animal microRNA genes". Cell Res. 18 (10): 985–96. doi:10.1038/cr.2008.278. PMC 2712117. PMID 18711447.CS1 maint: multiple names: authors list (link)
- Siepel A (October 2009). "Darwinian alchemy: Human genes from noncoding DNA". Genome Res. 19 (10): 1693–5. doi:10.1101/gr.098376.109. PMC 2765273. PMID 19797681.
- Darwin C (1859) On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life John Murray, London; modern reprint Charles Darwin; Julian Huxley (2003). The Origin of Species. Signet Classics. ISBN 978-0-451-52906-0. Published online at The complete work of Charles Darwin online: On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life.
- Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates. p. Glossary. ISBN 978-0-87893-189-7.
- Avers, Charlotte (1989). "Process and Pattern in Evolution". Oxford University Press. Cite journal requires
|journal=(help) - Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates. p. 320. ISBN 978-0-87893-189-7.
- Hahn, M.W. (2008). "Toward a selection theory of molecular evolution". Evolution. 62 (2): 255–265. doi:10.1111/j.1558-5646.2007.00308.x. PMID 18302709.
- Morjan C, Rieseberg L; Rieseberg (2004). "How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles". Mol. Ecol. 13 (6): 1341–56. doi:10.1111/j.1365-294X.2004.02164.x. PMC 2600545. PMID 15140081.
- Su H, Qu L, He K, Zhang Z, Wang J, Chen Z, Gu H; Qu; He; Zhang; Wang; Chen; Gu (2003). "The Great Wall of China: a physical barrier to gene flow?". Heredity. 90 (3): 212–9. doi:10.1038/sj.hdy.6800237. PMID 12634804.CS1 maint: multiple names: authors list (link)
- Short RV (1975). "The contribution of the mule to scientific thought". J. Reprod. Fertil. Suppl. (23): 359–64. PMID 1107543.
- Gross B, Rieseberg L; Rieseberg (2005). "The ecological genetics of homoploid hybrid speciation". J. Hered. 96 (3): 241–52. doi:10.1093/jhered/esi026. PMC 2517139. PMID 15618301.
- Burke JM, Arnold ML; Arnold (2001). "Genetics and the fitness of hybrids". Annu. Rev. Genet. 35: 31–52. doi:10.1146/annurev.genet.35.102401.085719. PMID 11700276.
- Vrijenhoek RC (2006). "Polyploid hybrids: multiple origins of a treefrog species". Curr. Biol. 16 (7): R245–7. doi:10.1016/j.cub.2006.03.005. PMID 16581499.
- Wendel J (2000). "Genome evolution in polyploids". Plant Mol. Biol. 42 (1): 225–49. doi:10.1023/A:1006392424384. PMID 10688139.
- Sémon M, Wolfe KH; Wolfe (2007). "Consequences of genome duplication". Current Opinion in Genetics & Development. 17 (6): 505–12. doi:10.1016/j.gde.2007.09.007. PMID 18006297.
- Comai L (2005). "The advantages and disadvantages of being polyploid". Nature Reviews Genetics. 6 (11): 836–46. doi:10.1038/nrg1711. PMID 16304599.
- Soltis P, Soltis D; Soltis (June 2000). "The role of genetic and genomic attributes in the success of polyploids". Proc. Natl. Acad. Sci. U.S.A. 97 (13): 7051–7. Bibcode:2000PNAS...97.7051S. doi:10.1073/pnas.97.13.7051. PMC 34383. PMID 10860970.
- Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF; Douady; Papke; Walsh; Boudreau; Nesbø; Case; Doolittle (2003). "Lateral gene transfer and the origins of prokaryotic groups". Annu Rev Genet. 37: 283–328. doi:10.1146/annurev.genet.37.050503.084247. PMID 14616063.CS1 maint: multiple names: authors list (link)
- Walsh T (2006). "Combinatorial genetic evolution of multiresistance". Current Opinion in Microbiology. 9 (5): 476–82. doi:10.1016/j.mib.2006.08.009. PMID 16942901.
- Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T; Nikoh; Ijichi; Shimada; Fukatsu (2002). "Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect". Proc. Natl. Acad. Sci. U.S.A. 99 (22): 14280–5. Bibcode:2002PNAS...9914280K. doi:10.1073/pnas.222228199. PMC 137875. PMID 12386340.CS1 maint: multiple names: authors list (link)
- Sprague G (1991). "Genetic exchange between kingdoms". Current Opinion in Genetics & Development. 1 (4): 530–3. doi:10.1016/S0959-437X(05)80203-5. PMID 1822285.
- Gladyshev EA, Meselson M, Arkhipova IR; Meselson; Arkhipova (May 2008). "Massive horizontal gene transfer in bdelloid rotifers". Science (Submitted manuscript). 320 (5880): 1210–3. Bibcode:2008Sci...320.1210G. doi:10.1126/science.1156407. PMID 18511688.CS1 maint: multiple names: authors list (link)
- Baldo A, McClure M; McClure (1 September 1999). "Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts". J. Virol. 73 (9): 7710–21. doi:10.1128/JVI.73.9.7710-7721.1999. PMC 104298. PMID 10438861.
- Poole A, Penny D; Penny (2007). "Evaluating hypotheses for the origin of eukaryotes". BioEssays. 29 (1): 74–84. doi:10.1002/bies.20516. PMID 17187354.
- Leavitt, Robert Botanical Gazette 1909 vol.47 no.1 January A Vegetative Mutant, and the Principle of Homoeosis in Plants https://www.jstor.org/pss/2466778
- edited by Scott, Eugenie C.; Branch, Glenn (2006). Not in our classrooms : why intelligent design is wrong for our schools (1st ed.). Boston: Beacon Press. p. 47. ISBN 978-0807032787.CS1 maint: extra text: authors list (link)
- "Young Earth Creationism". National Center for Science Education. 17 October 2008. Retrieved 18 May 2012.
- "Old Earth Creationism". National Center for Science Education. 17 October 2008. Retrieved 18 May 2012.
- Archived 26 January 2012 at the Wayback Machine, p. 12. American Association for the Advancement of Science
- Claim CB902: "Microevolution is distinct from macroevolution", TalkOrigins Archive
- "Archived copy". Archived from the original on 23 July 2011. Retrieved 29 July 2006.CS1 maint: archived copy as title (link)
- Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates.
- Complete sourced list of observed instances of speciation, TalkOrigins Archive
- Awbrey, Frank T. (1981). "Defining "Kinds" — Do Creationists Apply a Double Standard?". National Center for Science Education.
