Gene flow
In population genetics, gene flow (also known as gene migration or allele flow) is the transfer of genetic variation from one population to another. If the rate of gene flow is high enough, then two populations are considered to have equivalent allele frequencies and therefore effectively be a single population. It has been shown that it takes only "One migrant per generation" to prevent populations from diverging due to drift.[1] Gene flow is an important mechanism for transferring genetic diversity among populations. Migrants change the distribution of genetic diversity within the populations, by modifying the allele frequencies (the proportion of members carrying a particular variant of a gene). High rates of gene flow can reduce the genetic differentiation between the two groups, increasing homogeneity. For this reason, gene flow has been thought to constrain speciation by combining the gene pools of the groups, thus preventing the development of differences in genetic variation that would have led to full speciation.[2] In some cases migration may also result in the addition of novel genetic variants to the gene pool of a species or population.
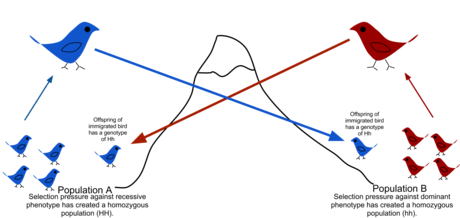
There are a number of factors that affect the rate of gene flow between different populations. Gene flow is expected to be lower in species that have low dispersal or mobility, that occur in fragmented habitats, where there is long distances between populations, and when there are small population sizes.[3][4] Mobility plays an important role in the migration rate, as highly mobile individuals tend to have greater migratory prospects. Although animals are thought to be more mobile than plants, pollen and seeds may be carried great distances by animals or wind. When gene flow is impeded, there can be an increase in inbreeding, measured by the inbreeding coefficient (F) within a population. For example, many island populations have low rates of gene flow due to geographic isolation and small population sizes. The Black Footed Rock Wallaby has several inbred populations that live on various islands off the coast of Australia. The population is so strongly isolated that lack of gene flow has led to high rates of inbreeding.[5]
Measuring gene flow
Decrease in population size leads to increased divergence due to drift, while migration reduces divergence and inbreeding. Gene flow can be measured by using the effective population size () and the net migration rate per generation (m). Using the approximation based on the Island model, the effect of migration can be calculated for a population in terms of the degree of genetic differentiation().[6] This formula accounts for the proportion of total molecular marker variation among populations, averaged over loci.[7] When there is one migrant per generation, the inbreeding coefficient () equals 0.2. However, when there is less than 1 migrant per generation (no migration), the inbreeding coefficient rises rapidly resulting in fixation and complete divergence ( = 1). The most common is < 0.25. This means there is some migration happening. Measures of population structure range from 0 to 1. When gene flow occurs via migration the deleterious effects of inbreeding can be ameliorated.[1]
The formula can be modified to solve for the migration rate when is known: , Nm = number of migrants [1].
Barriers to gene flow
Allopatric speciation
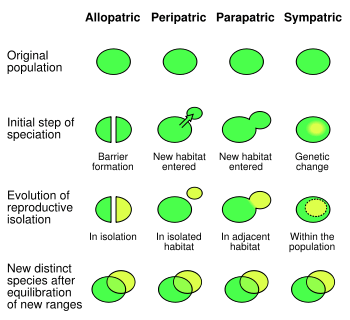
When gene flow is blocked by physical barriers, this results in Allopatric speciation or a geographical isolation that does not allow populations of the same species to exchange genetic material. Physical barriers to gene flow are usually, but not always, natural. They may include impassable mountain ranges, oceans, or vast deserts. In some cases, they can be artificial, man-made barriers, such as the Great Wall of China, which has hindered the gene flow of native plant populations.[8] One of these native plants, Ulmus pumila, demonstrated a lower prevalence of genetic differentiation than the plants Vitex negundo, Ziziphus jujuba, Heteropappus hispidus, and Prunus armeniaca whose habitat is located on the opposite side of the Great Wall of China where Ulmus pumila grows.[8] This is because Ulmus pumila has wind-pollination as its primary means of propagation and the latter-plants carry out pollination through insects.[8] Samples of the same species which grow on either side have been shown to have developed genetic differences, because there is little to no gene flow to provide recombination of the gene pools.
Sympatric speciation
Barriers to gene flow need not always be physical. Sympatric speciation happens when new species from the same ancestral species arise along the same range. This is often a result of a reproductive barrier. For example, two palm species of Howea found on Lord Howe Island were found to have substantially different flowering times correlated with soil preference, resulting in a reproductive barrier inhibiting gene flow.[9] Species can live in the same environment, yet show very limited gene flow due to reproductive barriers, fragmentation, specialist pollinators, or limited hybridization or hybridization yielding unfit hybrids. A cryptic species is a species that humans cannot tell is different without the use of genetics. Moreover, gene flow between hybrid and wild populations can result in loss of genetic diversity via genetic pollution, assortative mating and outbreeding. In human populations, genetic differentiation can also result from endogamy, due to differences in caste, ethnicity, customs and religion.
Human assisted gene-flow
Genetic rescue
Gene flow can also be used to assist species which are threatened with extinction. When a species exist in small populations there is an increased risk of inbreeding and greater susceptibility to loss of diversity due to drift. These populations can benefit greatly from the introduction of unrelated individuals who can increase diversity and reduce the amount of inbreeding, and thus increase overall fitness. This was demonstrated in the lab with two bottleneck strains of Drosophila melanogaster, in which crosses between the two populations reversed the effects of inbreeding and led to greater chances of survival in not only one generation but two.[10]
Genetic pollution
Human activities such as movement of species and modification of landscape can result in genetic pollution, hybridization, introgression and genetic swamping. These processes can lead to homogenization or replacement of local genotypes as a result of either a numerical and/or fitness advantage of introduced plant or animal.[11] Nonnative species can threaten native plants and animals with extinction by hybridization and introgression either through purposeful introduction by humans or through habitat modification, bringing previously isolated species into contact. These phenomena can be especially detrimental for rare species coming into contact with more abundant ones which can occur between island and mainland species. Interbreeding between the species can cause a 'swamping' of the rarer species' gene pool, creating hybrids that supplant the native stock. This is a direct result of evolutionary forces such as natural selection, as well as genetic drift, which lead to the increasing prevalence of advantageous traits and homogenization. The extent of this phenomenon is not always apparent from outward appearance alone. While some degree of gene flow occurs in the course of normal evolution, hybridization with or without introgression may threaten a rare species' existence.[12][13] For example, the Mallard is an abundant species of duck that interbreeds readily with a wide range of other ducks and poses a threat to the integrity of some species.[14]
Urbanization
There are two main models for how urbanization affects gene flow of urban populations. The first is through habitat fragmentation, also called urban fragmentation, in which alterations to the landscape that disrupt or fragment the habitat decrease genetic diversity. The second is called the urban facilitation model, and suggests that in some populations, gene flow is enabled by anthropogenic changes to the landscape. Urban facilitation of gene flow connects populations, reduces isolation, and increases gene flow into an area which would otherwise not have this specific genome composition.[15]
Urban facilitation can occur in many different ways, but most of the mechanisms include bringing previously separated species into contact, either directly or indirectly. Altering a habitat through urbanization will cause habitat fragmentation, but could also potentially disrupt barriers and create a pathway, or corridor, that can connect two formerly separated species. The effectiveness of this depends on individual species’ dispersal abilities and adaptiveness to different environments to use anthropogenic structures to travel. Human-driven climate change is another mechanism by which southern-dwelling animals might be forced northward towards cooler temperatures, where they could come into contact with other populations not previously in their range. More directly, humans are known to introduce non-native species into new environments, which could lead to hybridization of similar species.[16]
This urban facilitation model was tested on a human health pest, the Western black widow spider (Latrodectus hesperus). A study by Miles et. al collected genome-wide single nucleotide polymorphism variation data in urban and rural spider populations and found evidence for increased gene flow in urban Western black widow spiders compared to rural populations. In addition, the genome of these spiders was more similar across rural populations than it was for urban populations, suggesting increased diversity, and therefore adaptation, in the urban populations of the Western black widow spider. Phenotypically, urban spiders are larger, darker, and more aggressive, which could lead to increased survival in urban environments. These findings demonstrate support for urban facilitation, as these spiders are actually able to spread and diversify faster across urban environments than they would in a rural one. However, it is also an example of how urban facilitation, despite increasing gene flow, is not necessarily beneficial to an environment, as Western black widow spiders have highly toxic venom and therefore pose risks for human health.[17]
Another example of urban facilitation is that of migrating bobcats (Lynx rufus) in the northern US and southern Canada. A study by Marrote et. al sequenced fourteen different microsatellite loci in bobcats across the Great Lakes region, and found that longitude affected the interaction between anthropogenic landscape alterations and bobcat population gene flow. While rising global temperatures push bobcat populations into northern territory, increased human activity also enables bobcat migration northward. The increased human activity brings increased roads and traffic, but also increases road maintenance, plowing, and snow compaction, inadvertently clearing a path for bobcats to travel by. The anthropogenic influence on bobcat migration pathways is an example of urban facilitation via opening up a corridor for gene flow. However, in the bobcat’s southern range, an increase in roads and traffic is correlated with a decrease in forest cover, which hinders bobcat population gene flow through these areas. Somewhat ironically, the movement of bobcats northward is caused by human-driven global warming, but is also enabled by increased anthropogenic activity in northern ranges that make these habitats more suitable to bobcats.[18]
Consequences of urban facilitation vary from species to species. Positive effects of urban facilitation can occur when increased gene flow enables better adaptation and introduces beneficial alleles, and would ideally increase biodiversity. This has implications for conservation: for example, urban facilitation benefits an endangered species of tarantula and could help increase the population size. Negative effects would occur when increased gene flow is maladaptive and causes the loss of beneficial alleles. In the worst case scenario, this would lead to genomic extinction through a hybrid swarm. It is also important to note that in the scheme of overall ecosystem health and biodiversity, urban facilitation is not necessarily beneficial, and generally applies to urban adapter pests.[17] Examples of this include the previously mentioned Western black widow spider, and also the cane toad, which was able to use roads by which to travel and overpopulate Australia.[15]
Gene flow between species
Horizontal gene transfer
Horizontal gene transfer (HGT) refers to the transfer of genes between organisms in a manner other than traditional reproduction, either through transformation (direct uptake of genetic material by a cell from its surroundings), conjugation (transfer of genetic material between two bacterial cells in direct contact), transduction (injection of foreign DNA by a bacteriophage virus into the host cell) or GTA-mediated transduction (transfer by a virus-like element produced by a bacterium) .[19][20]
Viruses can transfer genes between species.[21] Bacteria can incorporate genes from dead bacteria, exchange genes with living bacteria, and can exchange plasmids across species boundaries.[22] "Sequence comparisons suggest recent horizontal transfer of many genes among diverse species including across the boundaries of phylogenetic 'domains'. Thus determining the phylogenetic history of a species can not be done conclusively by determining evolutionary trees for single genes."[23]
Biologist Gogarten suggests "the original metaphor of a tree no longer fits the data from recent genome research". Biologists [should] instead use the metaphor of a mosaic to describe the different histories combined in individual genomes and use the metaphor of an intertwined net to visualize the rich exchange and cooperative effects of horizontal gene transfer.[24]
"Using single genes as phylogenetic markers, it is difficult to trace organismal phylogeny in the presence of HGT. Combining the simple coalescence model of cladogenesis with rare HGT events suggest there was no single last common ancestor that contained all of the genes ancestral to those shared among the three domains of life. Each contemporary molecule has its own history and traces back to an individual molecule cenancestor. However, these molecular ancestors were likely to be present in different organisms at different times."[25]
Hybridization
In some instances, when a species has a sister species and breeding capabilities are possible due to the removal of previous barriers or through introduction due to human intervention, species can hybridize and exchange genes and corresponding traits. This exchange is not always clear-cut, for sometimes the hybrids may look identical to the original species phenotypically but upon testing the mtDNA it is apparent that hybridization has occurred. Differential hybridization also occurs because some traits and DNA are more readily exchanged than others, and this is a result of selective pressure or the absence thereof that allows for easier transaction. In instances in which the introduced species begins to replace the native species, the native species becomes threatened and the biodiversity is reduced, thus making this phenomenon negative rather than a positive case of gene flow that augments genetic diversity. Introgression is the replacement of the native species genes with that of the invader species. It is important to note that hybrids are generally deemed less "fit" than their parental generation, and as a result is a closely monitored genetic issue as the ultimate goal in conservation genetics is to maintain the genetic integrity of a species and preserve biodiversity.
Examples

While gene flow can greatly enhance the fitness of a population, it can also have negative consequences depending on the population and the environment in which they reside. The effects of gene flow are context-dependent.
- Fragmented Population: fragmented landscapes such as the Galapagos Islands are an ideal place for adaptive radiation to occur as a result of differing geography. Darwin's finches likely experienced allopatric speciation in some part due to differing geography, but that doesn't explain why we see so many different kinds of finches on the same island. This is due to adaptive radiation, or the evolution of varying traits in light of competition for resources. Gene flow moves in the direction of what resources are abundant at a given time.[26]
- Island Population: The marine iguana is an endemic species of the Galapagos Islands, but it evolved from a mainland ancestor of land iguana. Due to geographic isolation gene flow between the two species was limited and differing environments caused the marine iguana to evolve in order to adapt to the island environment. For instance, they are the only iguana that has evolved the ability to swim.
- Human Populations: Two theories exist for the human evolution throughout the world. The first is known as the multiregional model in which modern human variation is seen as a product of radiation of Homo erectus out of Africa after which local differentiation led to the establishment of regional population as we see them now.[27][28] Gene flow plays an important role in maintaining a grade of similarities and preventing speciation. In contrast the single origin theory assumes that there was a common ancestral population originating in Africa of Homo sapiens which already displayed the anatomical characteristics we see today. This theory minimizes the amount of parallel evolution that is needed.[28]
- Butterflies: Comparisons between sympatric and allopatric populations of Heliconius melpomene, H. cydno, and H. timareta revealed a genome-wide trend of increased shared variation in sympatry, indicative of pervasive interspecific gene flow.[29]
- Human-mediated gene flow: The captive genetic management of threatened species is the only way in which humans attempt to induce gene flow in ex situ situation. One example is the giant panda which is part of an international breeding program in which genetic materials are shared between zoological organizations in order to increase genetic diversity in the small populations. As a result of low reproductive success, artificial insemination with fresh/frozen-thawed sperm was developed which increased cub survival rate. A 2014 study found that high levels of genetic diversity and low levels of inbreeding were estimated in the breeding centers.[30]
- Plants: Two populations of monkeyflowers were found to use different pollinators (bees and hummingbirds) that limited gene flow, resulting in genetic isolation, eventually producing two different species, Mimulus lewisii and Mimulus cardinalis .[31]
- Sika deer: Sika deer were introduced into Western Europe, and they reproduce easily with the native red deer. This translocation of Sika deer has led to introgression and there are no longer "pure" red deer in the region, and all can be classified as hybrids.[32]
- Bobwhite quail: Bobwhite quail were translocated from the southern part of the United States to Ontario in order to increase population numbers and game for hunting.The hybrids that resulted from this translocation was less fit than the native population and were not adapted to survived the Northern Winters.[33]
References
- Frankham, Richard; Briscoe, David A.; Ballou, Jonathan D. (2002-03-14). Introduction to Conservation Genetics. Cambridge University Press. ISBN 9780521639859.
- Daniel I. Bolnick and Patrik Nosil (2007). "Natural Selection in Populations Subject to a Migration Load". Evolution. 61 (9): 2229–2243. doi:10.1111/j.1558-5646.2007.00179.x. PMID 17767592.
- Hastings, A; Harrison, S (November 1994). "Metapopulation Dynamics and Genetics". Annual Review of Ecology and Systematics. 25 (1): 167–188. doi:10.1146/annurev.es.25.110194.001123.
- "Effects of life history traits on genetic diversity in plant species". Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 351 (1345): 1291–1298. January 1997. doi:10.1098/rstb.1996.0112.
- Eldridge, Mark D. B.; King, Juliet M.; Loupis, Anne K.; Spencer, Peter B. S.; Taylor, Andrea C.; Pope, Lisa C.; Hall, Graham P. (June 1999). "Unprecedented Low Levels of Genetic Variation and Inbreeding Depression in an Island Population of the Black-Footed Rock-Wallaby". Conservation Biology. 13 (3): 531–541. doi:10.1046/j.1523-1739.1999.98115.x.
- Neigel, Joseph E. (1996). "Estimation of Effective Population Size and Migration Parameters From Genetic Data". In Smith, Thomas B.; Wayne, Robert K. (eds.). Molecular Genetic Approaches in Conservation. Oxford University Press. pp. 329–346. ISBN 978-0-19-534466-0.
- Rogers, D. L., & Montalvo, A. M. (2004). Genetically appropriate choices for plant materials to maintain biological diversity. University of California. Report to the USDA Forest Service, Rocky Mountain Region, Lakewood, CO. www. f s I ed. u s/ r, 2.
- Su H, Qu LJ, He K, Zhang Z, Wang J, Chen Z, Gu H (March 2003). "The Great Wall of China: a physical barrier to gene flow?". Heredity. 90 (3): 212–9. doi:10.1038/sj.hdy.6800237. PMID 12634804.
- Savolainen, Vincent; Anstett, Marie-Charlotte; Lexer, Christian; Hutton, Ian; Clarkson, James J.; Norup, Maria V.; Powell, Martyn P.; Springate, David; Salamin, Nicolas; Baker, William J. (8 February 2006). "Sympatric speciation in palms on an oceanic island". Nature. 441 (7090): 210–213. Bibcode:2006Natur.441..210S. doi:10.1038/nature04566. PMID 16467788.
- Heber, Sol; Briskie, James V.; Apiolaza, Luis A.; Skoulakis, Efthimios M. C. (13 August 2012). "A Test of the 'Genetic Rescue' Technique Using Bottlenecked Donor Populations of Drosophila melanogaster". PLOS ONE. 7 (8): e43113. Bibcode:2012PLoSO...743113H. doi:10.1371/journal.pone.0043113. PMC 3418252. PMID 22912802.
- Aubry, C.; Shoal, R.; Erickson, V. (2005). "Glossary". Grass cultivars: their origins, development, and use on national forests and grasslands in the Pacific Northwest. Corvallis, OR: USDA Forest Service; Native Seed Network (NSN), Institute for Applied Ecology. Archived from the original on 2006-02-22.
- Rhymer, Judith M.; Simberloff, Daniel (1996). "Extinction by Hybridization and Introgression". Annual Review of Ecology and Systematics. 27 (1): 83–109. doi:10.1146/annurev.ecolsys.27.1.83. JSTOR 2097230.
- Potts, Brad M.; Barbour, Robert C.; Hingston, Andrew B. (September 2001). Genetic Pollution from Farm Forestry using eucalypt species and hybrids; A report for the RIRDC/L&WA/FWPRDC; Joint Venture Agroforestry Program (PDF). RIRDC Publication No 01/114; RIRDC Project No CPF - 3A. Australian Government, Rural Industrial Research and Development Corporation. ISBN 978-0-642-58336-9. ISSN 1440-6845. Archived from the original (PDF) on 2004-01-02.
- "Hybrid Mallards-they're everywhere". Archived from the original on February 21, 2013. Retrieved January 23, 2013.
- Miles, Lindsay S.; Rivkin, L. Ruth; Johnson, Mark T. J.; Munshi-South, Jason; Verrelli, Brian C. (2019). "Gene flow and genetic drift in urban environments". Molecular Ecology. 28 (18): 4138–4151. doi:10.1111/mec.15221. PMID 31482608.
- Crispo, Erica; Moore, Jéan-Sebastian; Lee-Yaw, Julie A.; Gray, Suzanne M.; Haller, Benjamin C. (2011). "Broken barriers: Human‐induced changes to gene flow and introgression in animals". BioEssays. 33 (7): 508–518. doi:10.1002/bies.201000154. PMID 21523794.
- Miles, Lindsay J.; Johnson, J. Chadwick; Dyer, Rodney J.; Verrelli, Brian C. (2018). "Urbanization as a facilitator of gene flow in a human health pest". Molecule Ecology. 27 (16): 3219–3230. doi:10.1111/mec.14783. PMID 29972610.
- Marrottee, Robby R.; Bowman, Jeff; Wilson, Paul J. (2020). "Climate connectivity of the bobcat in the Great Lakes region". Ecology and Evolution. 10 (4): 2131–2144. doi:10.1002/ece3.6049. PMC 7042766. PMID 32128144.
- Johnston C, Martin B, Fichant G, Polard P, Claverys JP (March 2014). "Bacterial transformation: distribution, shared mechanisms and divergent control". Nature Reviews. Microbiology. 12 (3): 181–96. doi:10.1038/nrmicro3199. PMID 24509783.
- Lang, Andrew S.; Zhaxybayeva, Olga; Beatty, J. Thomas (11 June 2012). "Gene transfer agents: phage-like elements of genetic exchange". Nature Reviews Microbiology. 10 (7): 472–482. doi:10.1038/nrmicro2802. PMC 3626599. PMID 22683880.
- https://non.fiction.org/lj/community/ref_courses/3484/enmicro.pdf%5B%5D
- "Archived copy" (PDF). Archived from the original (PDF) on 2006-02-18. Retrieved 2005-12-31.CS1 maint: archived copy as title (link)
- "Archived copy". Archived from the original on 2005-10-16. Retrieved 2005-12-31.CS1 maint: archived copy as title (link)
- Horizontal Gene Transfer - A New Paradigm for Biology (from Evolutionary Theory Conference Summary), Esalen Center for Theory & Research
- "Archived copy". Archived from the original (PDF) on 2012-07-21. Retrieved 2005-12-31.CS1 maint: archived copy as title (link)
- Grant, Peter R.; Grant, B. Rosemary (2002-04-26). "Unpredictable Evolution in a 30-Year Study of Darwin's Finches". Science. 296 (5568): 707–711. Bibcode:2002Sci...296..707G. doi:10.1126/science.1070315. PMID 11976447. S2CID 36303685.
- Tobias, Phillip V.; Strong, Valerie; White, Heather (1985). Hominid Evolution: Past, Present, and Future : Proceedings of the Taung Diamond Jubilee International Symposium, Johannesburg and Mmabatho, Southern Africa, 27th January-4th February 1985. A.R. Liss. ISBN 978-0-8451-4202-8.
- Stringer, C.; Andrews, P (2 March 1988). "Genetic and fossil evidence for the origin of modern humans". Science. 239 (4845): 1263–1268. Bibcode:1988Sci...239.1263S. doi:10.1126/science.3125610. JSTOR 1700885. PMID 3125610.
- Martin, S. H.; Dasmahapatra, K. K.; Nadeau, N. J.; Salazar, C.; Walters, J. R.; Simpson, F.; Jiggins, C. D. (2013). "Genome-wide evidence for speciation with gene flow in heliconius butterflies". Genome Research. 23 (11): 1817–1828. doi:10.1101/gr.159426.113. PMC 3814882. PMID 24045163.
- Shan, Lei; Hu, Yibo; Zhu, Lifeng; Yan, Li; Wang, Chengdong; Li, Desheng; Jin, Xuelin; Zhang, Chenglin; Wei, Fuwen (2014-10-01). "Large-Scale Genetic Survey Provides Insights into the Captive Management and Reintroduction of Giant Pandas". Molecular Biology and Evolution. 31 (10): 2663–2671. doi:10.1093/molbev/msu210. PMID 25015646.
- Schemske, Douglas W.; Bradshaw, H. D. (1999-10-12). "Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus)". Proceedings of the National Academy of Sciences. 96 (21): 11910–11915. Bibcode:1999PNAS...9611910S. doi:10.1073/pnas.96.21.11910. PMC 18386. PMID 10518550.
- Storfer, Andrew. “Gene Flow and Endangered Species Translocations: a Topic Revisited.” Elsevier, Center for Ecology, Evolution and Behavior and T.H. Morgan School of Biological Sciences, May 1998.
- Storfer, Andrew (February 1999). "Gene flow and endangered species translocations: a topic revisited". Biological Conservation. 87 (2): 173–180. doi:10.1016/S0006-3207(98)00066-4.
External links
| Wikimedia Commons has media related to Gene flow. |
