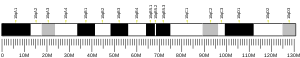
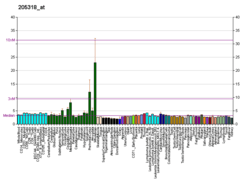
KIF5A
Kinesin heavy chain isoform 5A is a protein that in humans is encoded by the KIF5A gene.[5][6][7]
This gene encodes a member of the kinesin family of proteins. Members of this family are part of a multisubunit complex that functions as a microtubule motor in intracellular organelle transport. Mutations in this gene cause autosomal dominant spastic paraplegia 10.[7]
Clinical significance
Mutations in KIF5A were reported to cause hereditary spastic paraplegia type 10 (SPG1) by Douglas Marchuk in 2002.[10] In 2018, mutations in KIF5A were also found to cause amyotrophic lateral sclerosis by Bryan Traynor and John Landers.[11] KIF5A also plays a role in Alzheimer's disease by modulating the toxic effect of beta-amyloid on axonal transport of mitochondria.[12]
gollark: I an going to go to sleep soon. When I wake up after being unconscious for a bit, I still consider it me.
gollark: Sure!
gollark: "We scan your brain structure while it's not running/very fast and emulate it on a computer" is simple enough.
gollark: I don't see why you still insist on this version...
gollark: I still think that unless some fundamental things about consciousness which that assumes are figured out, and perhaps even then, this is kind of wasteful and useless.
References
- GRCh38: Ensembl release 89: ENSG00000155980 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000074657 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hamlin PJ, Jones PF, Leek JP, Bransfield K, Lench NJ, Aldersley MA, et al. (February 1999). "Assignment of GALGT encoding beta-1, 4N-acetylgalactosaminyl-transferase (GalNAc-T) and KIF5A encoding neuronal kinesin (D12S1889) to human chromosome band 12q13 by assignment to ICI YAC 26EG10 and in situ hybridization. medjph@stjames.leeds.ac.uk". Cytogenetics and Cell Genetics. 82 (3–4): 267–8. doi:10.1159/000015115. PMID 9858832.
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC (September 1999). "A new locus for autosomal dominant "pure" hereditary spastic paraplegia mapping to chromosome 12q13, and evidence for further genetic heterogeneity". American Journal of Human Genetics. 65 (3): 757–63. doi:10.1086/302555. PMC 1377983. PMID 10441583.
- "Entrez Gene: KIF5A kinesin family member 5A".
- Rahman A, Friedman DS, Goldstein LS (June 1998). "Two kinesin light chain genes in mice. Identification and characterization of the encoded proteins". The Journal of Biological Chemistry. 273 (25): 15395–403. doi:10.1074/jbc.273.25.15395. PMID 9624122.
- Rahman A, Kamal A, Roberts EA, Goldstein LS (September 1999). "Defective kinesin heavy chain behavior in mouse kinesin light chain mutants". The Journal of Cell Biology. 146 (6): 1277–88. doi:10.1083/jcb.146.6.1277. PMC 2156125. PMID 10491391.
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, et al. (November 2002). "A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10)". American Journal of Human Genetics. 71 (5): 1189–94. doi:10.1086/344210. PMC 385095. PMID 12355402.
- Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, et al. (March 2018). "Genome-wide Analyses Identify KIF5A as a Novel ALS Gene". Neuron. 97 (6): 1268–1283.e6. doi:10.1016/j.neuron.2018.02.027. PMC 5867896. PMID 29566793.
- Wang Q, Tian J, Chen H, Du H, Guo L (July 2019). "Amyloid beta-mediated KIF5A deficiency disrupts anterograde axonal mitochondrial movement". Neurobiology of Disease. 127: 410–418. doi:10.1016/j.nbd.2019.03.021. PMID 30923004.
Further reading
- Fichera M, Lo Giudice M, Falco M, Sturnio M, Amata S, Calabrese O, et al. (September 2004). "Evidence of kinesin heavy chain (KIF5A) involvement in pure hereditary spastic paraplegia". Neurology. 63 (6): 1108–10. doi:10.1212/01.wnl.0000138731.60693.d2. PMID 15452312.
- Niclas J, Navone F, Hom-Booher N, Vale RD (May 1994). "Cloning and localization of a conventional kinesin motor expressed exclusively in neurons". Neuron. 12 (5): 1059–72. doi:10.1016/0896-6273(94)90314-X. PMID 7514426.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Rahman A, Friedman DS, Goldstein LS (June 1998). "Two kinesin light chain genes in mice. Identification and characterization of the encoded proteins". The Journal of Biological Chemistry. 273 (25): 15395–403. doi:10.1074/jbc.273.25.15395. PMID 9624122.
- Rahman A, Kamal A, Roberts EA, Goldstein LS (September 1999). "Defective kinesin heavy chain behavior in mouse kinesin light chain mutants". The Journal of Cell Biology. 146 (6): 1277–88. doi:10.1083/jcb.146.6.1277. PMC 2156125. PMID 10491391.
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N (September 2000). "KIF5C, a novel neuronal kinesin enriched in motor neurons". The Journal of Neuroscience. 20 (17): 6374–84. doi:10.1523/JNEUROSCI.20-17-06374.2000. PMC 6772948. PMID 10964943.
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N (May 2002). "Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites". Nature. 417 (6884): 83–7. doi:10.1038/nature743. PMID 11986669.
- Macioce P, Gambara G, Bernassola M, Gaddini L, Torreri P, Macchia G, et al. (December 2003). "Beta-dystrobrevin interacts directly with kinesin heavy chain in brain". Journal of Cell Science. 116 (Pt 23): 4847–56. doi:10.1242/jcs.00805. PMID 14600269.
- Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, et al. (July 2004). "Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding". Genes & Development. 18 (14): 1737–52. doi:10.1101/gad.294904. PMC 478194. PMID 15256501.
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. (September 2005). "A human protein-protein interaction network: a resource for annotating the proteome". Cell. 122 (6): 957–68. doi:10.1016/j.cell.2005.08.029. hdl:11858/00-001M-0000-0010-8592-0. PMID 16169070.
- Blair MA, Ma S, Hedera P (March 2006). "Mutation in KIF5A can also cause adult-onset hereditary spastic paraplegia". Neurogenetics. 7 (1): 47–50. doi:10.1007/s10048-005-0027-8. PMID 16489470.
External links
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.