Denisovan
The Denisovans or Denisova hominins ( /dɪˈniːsəvə/ di-NEE-sə-və) are an extinct species or subspecies of archaic human that ranged across Asia during the Lower and Middle Paleolithic (potentially surviving as late as 30,000–14,500 years ago in New Guinea). Denisovans are known from few remains, and, consequently, most of what is known about them comes from DNA evidence. Pending consensus on their taxonomic status, they have been referred to as Homo denisova, H. altaiensis, or H. sapiens denisova.

The first Denisovan individual was identified in 2010 based on mitochondrial DNA (mtDNA) extracted from a juvenile female finger bone from the Siberian Denisova Cave. Nuclear DNA (nDNA) indicates close affinities with Neanderthals. The cave was also periodically inhabited by Neanderthals, but it is unclear whether they ever cohabited in the cave. Additional specimens from Denisova Cave were subsequently discovered, as was a single specimen from the Baishiya Karst Cave on the Tibetan Plateau in China. This indicates they lived in a wide variety of habitats, including forests, tundras, mountains and jungles. DNA evidence suggests they had dark skin, eyes, and hair, and had a Neanderthal-like build and facial features. However, they had larger molars which are reminiscent of Middle to Late Pleistocene archaic humans and australopithecines.
Denisovans apparently interbred with modern humans, with about 3–5% of the DNA of Melanesians and Aboriginal Australians and around 6% in Papuans deriving from Denisovans. Denisovans may have interbred with modern humans in New Guinea as recently as 15,000 years ago. There is also evidence of interbreeding with the local Neanderthal population, with about 17% of the Denisovan genome from Denisova Cave deriving from them. A first generation hybrid nicknamed "Denny" was discovered with a Denisovan father and a Neanderthal mother. 4% of the Denisovan genome comes from an unknown archaic human species which diverged from modern humans over one million years ago.
Taxonomy
It is debated whether Denisovans represent a distinct species of Homo or are an archaic subspecies of H. sapiens. DNA analyses showing Denisovans as a sister taxon of Neanderthals also concerns the classification of the latter as H. neanderthalensis or H. s. neanderthalensis. Proposed species names for Denisovans are H. denisova[1] or H. altaiensis.[2]
Discovery

Denisova Cave is in south-central Siberia, Russia in the Altai Mountains near the border with Kazakhstan, China and Mongolia. It is named after Denis (Dyonisiy), a Russian hermit who lived there in the 18th century. The cave was originally explored in the 1970s by Russian paleontologist Nikolai Ovodov, who was looking for remains of canids.[3]
In 2008, Michael Shunkov from the Russian Academy of Sciences and other Russian archaeologists from the Institute of Archaeology and Ethnology of Novosibirsk investigated the cave and found the finger bone of a juvenile female hominin originally dated to 50–30,000 years ago.[4][5] The estimate has changed to 76,200–51,600 years ago.[6] The specimen was originally named X-woman because matrilineal mitochondrial DNA (mtDNA) extracted from the bone demonstrated it to belong to a novel ancient hominin, genetically distinct from contemporary modern humans and Neanderthals.[4]
In 2019, Greek archaeologist Katerina Douka and colleagues radiocarbon dated specimens from Denisova Cave, and estimated that Denisova 2 (the oldest specimen) lived 195,000-122,700 years ago.[6] Older Denisovan DNA collected from sediments in the East Chamber dates to 217,000 years ago. Based on artifacts also discovered in the cave, hominin occupation (most likely by Denisovans) began 287±41 or 203±14 ka. Neanderthals were also present 193±12 ka and 97±11 ka, possibly concurrently with Denisovans.[7]
Specimens
The fossils of five distinct Denisovans from Denisova Cave have been identified through their Ancient DNA (aDNA): Denisova 2, Denisova 3, Denisova 4, Denisova 8, and Denisova 13. Denisova 11 was an F1 Denisovan-Neanderthal hybrid.[8] An mtDNA-based phylogenetic analysis of these individuals suggests that Denisova 2 is the oldest, followed by Denisova 8, while Denisova 3 and Denisova 4 were roughly contemporaneous.[9] During DNA sequencing, a low proportion of the Denisova 2, Denisova 4 and Denisova 8 genomes were found to have survived, but a high proportion of the Denisova 3 genome was intact.[9][10] Denisova 3 was cut into two, and the initial DNA sequencing of one fragment was later independently confirmed by sequencing the mtDNA from the second.[11]
These specimens remained the only known examples of Denisovans until 2019 when a research group led by Fahu Chen, Dongju Zhang and Jean-Jacques Hublin described a partial mandible discovered in 1980 by a Buddhist monk in the Baishiya Karst Cave on the Tibetan Plateau in China. The fossil became part of the collection of Lanzhou University, where it remained unstudied until 2010.[12] It was determined by ancient protein analysis to contain collagen that by sequence was found to have close affiliation to that of the Denisovans from Denisova Cave, while uranium decay dating of the carbonate crust enshrouding the specimen indicated it was more than 160,000 years old.[13]
Some older findings may or may not belong to the Denisovan line, but Asia is not well mapped in regards to human evolution. Such findings include the Dali skull,[14] the Xujiayao hominin,[15] Maba Man, the Jinniushan hominin, and the Narmada hominin.[16] The Xiahe mandible shows morphological similarities to some later East Asian fossils such as Penghu 1,[13][17] but also to Chinese H. erectus.[11]
| Name | Fossil elements | Age | Discovery | Place | Sex and age | Publication | Image | GenBank accession |
|---|---|---|---|---|---|---|---|---|
| Denisova 3 (aka X Woman)[18][11][4] | Fifth distal finger phalanx | 76.2–51.6 ka[6] | 2008 | Denisova cave (Russia) | 13.5 year adolescent female | 2010 |  Replica of part of the phalanx. | NC013993 |
| Denisova 4[18][14] | Permanent upper 2nd or 3rd molar | 84.1–55.2 ka[6] | 2000 | Denisova cave (Russia) | Adult male | 2010 |  Replica of the molar of Denisova. Part of the roots was destroyed to study the mtDNA. Their size and shape indicate it is neither neanderthal nor sapiens. |
FR695060 |
| Denisova 8[10] | Permanent upper 3rd molar | 136.4–105.6 ka[6] | 2010 | Denisova cave (Russia) | Adult male | 2015 | KT780370 | |
| Denisova 2[9] | Deciduous 2nd lower molar | 194.4–122.7 ka[6] | 1984 | Denisova cave (Russia) | Adolescent female | 2017 | KX663333 | |
| Xiahe mandible[13] | Partial mandible | > 160 ka | 1980 | Baishiya Cave (China) | 2019 |  |
||
| Denisova 11 (aka Denny, Denisovan x Neanderthal hybrid) [19] |
Arm or leg bone fragment | 118.1–79.3 ka[6] | 2012 | Denisova cave (Russia) | 13 year old adolescent female | 2016 |  |
|
| Denisova 13[20] | Parietal bone fragment | Found in layer 22[20] which dates to ~285±39 ka[7] | 2019 | Denisova cave (Russia) | pending |
Divergence times
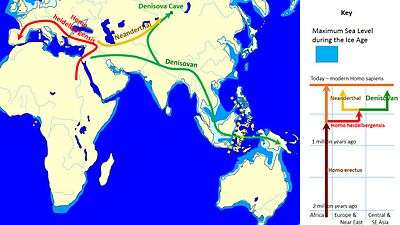
Sequenced mitochondrial DNA (mtDNA), preserved by the cool climate of the cave (average temperature is at freezing point), was extracted from Denisova 3 by a team of scientists led by Johannes Krause and Svante Pääbo from the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany. Denisova 3's mtDNA differs from that of modern humans by 385 bases (nucleotides) out of approximately 16,500, whereas the difference between modern humans and Neanderthals is around 202 bases. In comparison, the difference between chimpanzees and modern humans is approximately 1,462 mtDNA base pairs. This suggested that Denisovan mtDNA diverged from that of modern humans and Neanderthals about 1,313,500–779,300 years ago; whereas modern human and Neanderthal mtDNA diverged 618–321,200 years ago. Krause and colleagues then concluded that Denisovans were the descendants of an earlier migration of H. erectus out of Africa, completely distinct from modern humans and Neanderthals.[4]
However, according to the nuclear DNA (nDNA) of Denisova 3—which had an unusual degree of DNA preservation with only low-level contamination—Denisovans and Neanderthals were more closely related to each other than they were to modern humans. Using the percent distance from human–chimpanzee last common ancestor, Denisovans/Neanderthals split from modern humans about 804,000 years ago, and from each other 640,000 years ago.[18] Using a mutation rate of 1x10−9 or 0.5x10−9 per base pair (bp) per year, the Neanderthal/Denisovan split occurred around either 236–190,000 or 473–381,000 years ago respectively.[21] Using 1.1x10−8 per generation with a new generation every 29 years, the time is 744,000 years ago. Using 5x10−10 nucleotide site per year, it is 616,000 years ago. Using the latter dates, the split had likely already occurred by the time hominins spread out across Europe.[22] H. heidelbergensis is typically considered to have been the direct ancestor of Denisovans and Neanderthals, and sometimes also modern humans.[23] Due to the strong divergence in dental anatomy, they may have split before characteristic Neanderthal dentition evolved about 300,000 years ago.[18]
The more divergent Denisovan mtDNA has been interpreted as evidence of admixture between Denisovans and an unknown archaic human population,[24] possibly a relict H. erectus or H. erectus-like population about 53,000 years ago.[21] Alternatively, divergent mtDNA could have also resulted from the persistence of an ancient mtDNA lineage which only went extinct in modern humans and Neanderthals through genetic drift.[18] Modern humans contributed mtDNA to the Neanderthal lineage, but not to the Denisovan mitochondrial genomes yet sequenced.[25][26][27][28] The mtDNA sequence from the femur of a 400,000-year-old H. heidelbergensis from the Sima de los Huesos Cave in Spain was found to be related to those of Neanderthals and Denisovans, but closer to Denisovans,[29][30] and the authors posited that this mtDNA represents an archaic sequence which was subsequently lost in Neanderthals due to replacement by a modern-human-related sequence.[31]
Demographics
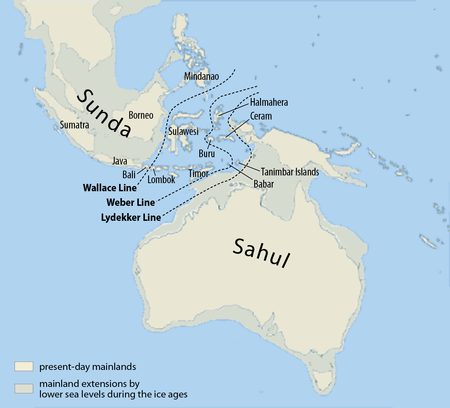
Though their remains have been identified in only two locations, traces of Denisovan DNA in modern humans suggest they ranged across East Asia,[32][33] and potentially western Eurasia.[34] In 2019, geneticist Guy Jacobs identified three distinct populations of Denisovans in, respectively, Siberia and East Asia, New Guinea and nearby islands, and Oceania and to a lesser extent across Asia. Using coalescent modeling, the Denisova Cave Denisovans split from the second population about 283,000 years ago; and from the third population about 363,000 years ago. This indicates that there was marked reproductive separation between Denisovan populations.[35] Based on the modern distribution of Denisovan DNA, Denisovans may have crossed the Wallace Line into Wallacea and also Sahul (New Guinea and Australia), with little back-migration to west of the line.[16] These Denisovans may have needed to cross large bodies of water.[35]
Using exponential distribution analysis on haplotype lengths, Jacobs calculated introgression into modern humans occurred about 29,900 years ago with the second population; and 45,700 years ago in the third population. Such a late date for the second population could indicate survival as late as 14,500 years ago, which would make them the latest surviving archaic human species. New Guineans have introgression from these two latter populations. A third wave appears to have introgressed into East Asia, but there is not enough DNA evidence to pinpoint a solid timeframe.[35]
The mtDNA from Denisova 4 bore a high similarity to that of Denisova 3, indicating that they belonged to the same population.[18] The genetic diversity among the Denisovans from Denisova Cave is on the lower range of what is seen in modern humans, and is comparable to that of Neanderthals. However, it is possible that the inhabitants of Denisova Cave were more or less reproductively isolated from other Denisovans, and that, across their entire range, Denisovan genetic diversity may have been much higher.[9]
Denisova Cave, over time of inhabitance, continually swung from a fairly warm and moderately humid pine and birch forest to a tundra or forest–tundra landscape.[7] Conversely, Baishiya Karst Cave is situated at a high elevation, an area characterized by low temperature, low oxygen, and poor resource availability. Colonization of high-altitude regions, due to such harsh conditions, was previously assumed to have only been accomplished by modern humans.[13] Denisovans seem to have also inhabited the jungles of Southeast Asia.[33]
Anatomy
Little is known of the precise anatomical features of the Denisovans since the only physical remains discovered so far are a finger bone, three teeth, long bone fragments, a partial jawbone,[12] and a parietal bone skull fragment.[20] The finger bone is within the modern human range of variation for women,[11] which is in contrast to the large, robust molars which are more similar to those of Middle to Late Pleistocene archaic humans. The third molar is outside of the range of any Homo species except H. habilis and H. rudolfensis, and is more like those of australopithecines. The second molar is larger than those of modern humans and Neanderthals, and is more similar to those of H. erectus and H. habilis.[18] Like Neanderthals, the mandible had a gap behind the molars, and the front teeth were flattened; but Denisovans lacked a high mandibular body, and the mandibular symphysis at the midline of the jaw was more receding.[13][17] The parietal is reminiscent of that of H. erectus.[36]
A facial reconstruction has been generated by comparing methylation at individual genetic loci associated with facial structure.[37] This analysis suggested that Denisovans, much like Neanderthals, had a long, broad, and projecting face; larger nose; sloping forehead; protruding jaw; elongated and flattened skull; and wide chest and hips. However, the Denisovan tooth row was longer than that of Neanderthals and anatomically modern humans.[38]
The Denisovan genome from Denisova Cave has variants of genes which, in modern humans, are associated with dark skin, brown hair, and brown eyes.[39] The Denisovan genome also contains a variant region around the EPAS1 gene that in Tibetans assists with adaptation to low oxygen levels at high elevation,[40][13] and in a region containing the WARS2 and TBX15 loci which affect body-fat distribution in the Inuit.[41] In Papuans, introgressed Neanderthal alleles are highest in frequency in genes expressed in the brain, whereas Denisovan alleles have highest frequency in genes expressed in bones and other tissue.[42]
Artifacts


Early Middle Paleolithic stone tools from Denisova Cave were characterized by discoidal (disk-like) cores and Kombewa cores, but Levallois cores and flakes were also present. There were scrapers, denticulate tools, and notched tools, deposited about 287±41 thousand years ago in the Main Chamber of the cave; and about 269±97 thousand years ago in the South Chamber; up to 170±19 thousand and 187±14 thousand years ago in the Main and East Chambers, respectively.[7]
Middle Middle Paleolithic assemblages were dominated by flat, discoidal, and Levallois cores, and there were some isolated sub-prismatic cores. There were predominantly side scrapers (a scraper with only the sides used to scrape), but also notched-denticulate tools, end-scrapers (a scraper with only the ends used to scrape), burins, chisel-like tools, and truncated flakes. These dated to 156±15 thousand years ago in the Main Chamber, 58±6 thousand years ago in the East Chamber, and 136±26–47±8 thousand years ago in the South Chamber.[7]
Early Upper Paleolithic artifacts date to 44±5 thousand years ago in the Main Chamber, 63±6 thousand years ago in the East Chamber, and 47±8 thousand years ago in the South Chamber, though some layers of the East Chamber seem to have been disturbed. There was blade production and Levallois production, but scrapers were again predominant. A well-developed, Upper Paleolithic stone bladelet technology distinct from the previous scrapers began accumulating in the Main Chamber around 36±4 thousand years ago.[7]
In the Upper Paleolithic layers, there were also several bone tools and ornaments: a marble ring, an ivory ring, an ivory pendant, a red deer tooth pendant, an elk tooth pendant, a chloritolite bracelet, and a bone needle. However, Denisovans are only confirmed to have inhabited the cave until 55 ka; the dating of Upper Paleolithic artifacts overlaps with modern human migration into Siberia (though there are no occurrences in the Altai region); and the DNA of the only specimen in the cave dating to the time interval (Denisova 14) is too degraded to confirm a species identity, so the attribution of these artifacts is unclear.[43][7]
Interbreeding
Analyses of modern humans genomes show past interbreeding with at least two groups of archaic humans, Neanderthals[44] and Denisovans,[18][45] and that such interbreeding events occurred on multiple occasions. Comparisons of the Denisovan, Neanderthal, and modern human genomes has revealed evidence for a complex web of interbreeding among these lineages.[44]
Archaic humans
As much as 17% of the Denisovan genome from Denisova Cave represents DNA from the local Neanderthal population.[44] The Denisovan genome shares more derived alleles with the Altai Neanderthal genome from Siberia than with the Vindija Cave Neanderthal genome from Croatia or the Mezmaiskaya cave Neanderthal genome from the Caucasus, suggesting that the gene flow came from a population that was more closely related to the Altai Neanderthal.[46] However, Denny's Denisovan father had the typical Altai Neanderthal introgression, while her Neanderthal mother represented a population more closely related to Vindija Neanderthals than to those of Altai.[47]
About 4% of the Denisovan genome derives from an unidentified archaic hominin,[44] perhaps the source of the anomalous ancient mtDNA, indicating this species diverged from Neanderthals and humans over a million years ago. The only identified Homo species of Late Pleistocene Asia are H. erectus and H. heidelbergensis.[46][48] It is unclear if Denisovan populations which introgressed into modern humans had this archaic hominin ancestry.[35]
Before splitting from Neanderthals, their ancestors ("Neandersovans") migrating out of Africa into Europe apparently interbred with an unidentified "superarchaic" human species who were already present there; these superarchaics were the descendants of a very early migration out of Africa around 1.9 mya.[49]
Modern humans
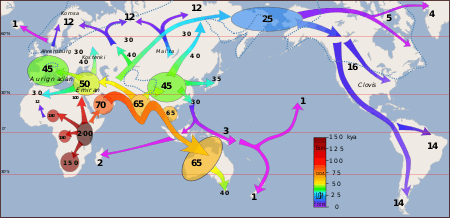
A 2011 study found that Denisovan DNA is prevalent in Australian Aborigines, Near Oceanians, Polynesians, Fijians, East Indonesians and Mamanwans (from the Philippines); but not in East Asians, western Indonesians, Jahai people (from Malaysia) or Onge (from the Andaman Islands). This means that Denisovan introgression occurred within the Pacific region rather than on the Asian mainland, and that ancestors of the latter groups were not present in Southeast Asia at the time, which in turn means that eastern Asia was settled by modern humans in two distinct migrations.[33] In the Melanesian genome, about 4–6%[18] or 1.9–3.4% derives from Denisovan introgression.[50] New Guineans and Australian Aborigines have the most introgressed DNA,[16] but Aborigines have less than New Guineans.[51] In Papuans, less Denisovan ancestry is seen in the X chromosome than autosomes, and some autosomes (such as chromosome 11) also have less Denisovan ancestry, which could indicate hybrid incompatibility. The former observation could also be explained by less female Denisovan introgression into modern humans, or more female modern human immigrants who diluted Denisovan X chromosome ancestry.[39]
In contrast, 0.2% derives from Denisovan ancestry in mainland Asians and Native Americans.[52] South Asians were found to have levels of Denisovan admixture similar to that seen in East Asians.[53] The discovery of the 40,000 year old Chinese modern human Tianyuan Man lacking Denisovan DNA significantly different from the levels in modern-day East Asians discounts the hypothesis that immigrating modern humans simply diluted Denisovan ancestry whereas Melanesians lived in reproductive isolation.[54][16] A 2018 study of Han Chinese, Japanese, and Dai genomes showed that modern East Asians have DNA from two different Denisovan populations: one similar to the Denisovan DNA found in Papuan genomes, and a second that is closer to the Denisovan genome from Denisova Cave. This could indicate two separate introgression events involving two different Denisovan populations. In South Asian genomes, DNA only came from the same single Denisovan introgression seen in Papuans.[53] A 2019 study found a third wave of Denisovans which introgressed into East Asians. Introgression, also, may not have immediately occurred when modern humans immigrated into the region.[35]
In other regions of the world, archaic introgression into humans stems from a group of Neanderthals related to those which inhabited Vindija Cave, Croatia, as opposed to archaics related to Siberian Neanderthals and Denisovans. However, about 13.1 and 3.3% of the archaic DNA in the modern Icelandic genome descends from these two latter groups, respectively, and such a high percentage could indicate a western Eurasian population of Denisovans which introgressed into either Vindija-related Neanderthals or immigrating modern humans.[34]
Denisovan genes may have helped early modern humans migrating out of Africa to acclimatize. Although not present in the sequenced Denisovan genome, the distribution pattern and divergence of HLA-B*73 from other HLA alleles (involved in the immune system's natural killer cell receptors) has led to the suggestion that it introgressed from Denisovans into modern humans in West Asia. In a 2011 study, half of the HLA alleles of modern Eurasians were shown to represent archaic HLA haplotypes, and were inferred to be of Denisovan or Neanderthal origin.[55] A haplotype of EPAS1, likely introgressed into Tibetans from Denisovans, allows them to live at high elevations in a low-oxygen environment.[40][13] Genes related to phospholipid transporters (which are involved in fat metabolism) and to trace amine-associated receptors (involved in smelling) are more active in people with more Denisovan ancestry.[56]
See also
- Timeline of human evolution – Chronological outline of major events in the development of the human species
- Red Deer Cave people – Archaic humans from 12,500 BCE in southwest China
- Homo luzonensis – Archaic human from Luzon, Philippines
- Homo naledi – Small-brained South African archaic human
- Homo floresiensis – Archaic human from Flores, Indonesia
References
- Douglas, M. M.; Douglas, J. M. (2016). Exploring Human Biology in the Laboratory. Morton Publishing Company. p. 324. ISBN 9781617313905.
- Zubova, A.; Chikisheva, T.; Shunkov, M. V. (2017). "The Morphology of Permanent Molars from the Paleolithic Layers of Denisova Cave". Archaeology, Ethnology & Anthropology of Eurasia. 45: 121–134. doi:10.17746/1563-0110.2017.45.1.121-134.
- Ovodov, N. D.; Crockford, S. J.; Kuzmin, Y. V.; Higham, T. F.; Hodgins, G. W.; van der Plicht, J. (2011). "A 33,000-Year-Old Incipient Dog from the Altai Mountains of Siberia: Evidence of the Earliest Domestication Disrupted by the Last Glacial Maximum". PLOS ONE. 6 (7): e22821. Bibcode:2011PLoSO...622821O. doi:10.1371/journal.pone.0022821. PMC 3145761. PMID 21829526.
- Krause, J.; Fu, Q.; Good, J. M.; Viola, B.; Shunkov, M. V.; Derevianko, A. P. & Pääbo, S. (2010). "The complete mitochondrial DNA genome of an unknown hominin from southern Siberia". Nature. 464 (7290): 894–897. Bibcode:2010Natur.464..894K. doi:10.1038/nature08976. PMID 20336068.
- Reich, D. (2018). Who We Are and How We Got Here. Oxford University Press. p. 53. ISBN 978-0-19-882125-0.
- Douka, K. (2019). "Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave". Nature. 565 (7741): 640–644. Bibcode:2019Natur.565..640D. doi:10.1038/s41586-018-0870-z. PMID 30700871.
- Jacobs, Zenobia; Li, Bo; Shunkov, Michael V.; Kozlikin, Maxim B.; Bolikhovskaya, Nataliya S.; Agadjanian, Alexander K.; Uliyanov, Vladimir A.; Vasiliev, Sergei K.; O’Gorman, Kieran; Derevianko, Anatoly P.; Roberts, Richard G. (January 2019). "Timing of archaic hominin occupation of Denisova Cave in southern Siberia". Nature. 565 (7741): 594–599. Bibcode:2019Natur.565..594J. doi:10.1038/s41586-018-0843-2. ISSN 1476-4687. PMID 30700870.
- Warren, M. (2018). "Mum's a Neanderthal, Dad's a Denisovan: First discovery of an ancient-human hybrid – Genetic analysis uncovers a direct descendant of two different groups of early humans". Nature. 560 (7719): 417–418. Bibcode:2018Natur.560..417W. doi:10.1038/d41586-018-06004-0. PMID 30135540.
- Slon, V.; Viola, B.; Renaud, G.; Gansauge, M.-T.; Benazzi, S.; Sawyer, S.; Hublin, J.-J.; Shunkov, M. V.; Derevianko, A. P.; Kelso, J.; Prüfer, K.; Meyer, M.; Pääbo, S. (2017). "A fourth Denisovan individual". Science Advances. 3 (7): e1700186. Bibcode:2017SciA....3E0186S. doi:10.1126/sciadv.1700186. PMC 5501502. PMID 28695206.
- Sawyer, S.; Renaud, G.; Viola, B.; Hublin, J.-J.; Gansauge, M.-T.; Shunkov, M. V.; Derevianko, A. P.; Prüfer, K.; Kelso, J.; Pääbo, S. (2015). "Nuclear and mitochondrial DNA sequences from two Denisovan individuals". Proceedings of the National Academy of Sciences. 112 (51): 15696–700. Bibcode:2015PNAS..11215696S. doi:10.1073/pnas.1519905112. PMC 4697428. PMID 26630009.
- Bennett, E. A.; Crevecoeur, I.; Viola, B.; et al. (2019). "Morphology of the Denisovan phalanx closer to modern humans than to Neanderthals". Science Advances. 5 (9): eaaw3950. Bibcode:2019SciA....5.3950B. doi:10.1126/sciadv.aaw3950. PMC 6726440. PMID 31517046.
- Gibbons, A. (2019). "First fossil jaw of Denisovans finally puts a face on elusive human relatives". Science. doi:10.1126/science.aax8845.
- Chen, F.; Welker, F.; Shen, C.-C.; et al. (2019). "A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau" (PDF). Nature. 569 (7756): 409–412. Bibcode:2019Natur.569..409C. doi:10.1038/s41586-019-1139-x. PMID 31043746.
- Callaway, Ewen (2010). "Fossil genome reveals ancestral link". Nature. 468 (7327): 1012. Bibcode:2010Natur.468.1012C. doi:10.1038/4681012a. PMID 21179140.
- Ao, H.; Liu, C.-R.; Roberts, A. P. (2017). "An updated age for the Xujiayao hominin from the Nihewan Basin, North China: Implications for Middle Pleistocene human evolution in East Asia". Journal of Human Evolution. 106: 54–65. Bibcode:2017AGUFMPP13B1080A. doi:10.1016/j.jhevol.2017.01.01 (inactive 29 May 2020). PMID 28434540.
- Cooper, A.; Stringer, C. B. (2013). "Did the Denisovans Cross Wallace's Line?". Science. 342 (6156): 321–23. Bibcode:2013Sci...342..321C. doi:10.1126/science.1244869. PMID 24136958.
- Warren, M. (2019). "Biggest Denisovan fossil yet spills ancient human's secrets". Nature News. 569 (7754): 16–17. Bibcode:2019Natur.569...16W. doi:10.1038/d41586-019-01395-0. PMID 31043736.
- Reich, D.; Green, R. E.; Kircher, M.; et al. (2010). "Genetic history of an archaic hominin group from Denisova Cave in Siberia" (PDF). Nature. 468 (7327): 1053–60. Bibcode:2010Natur.468.1053R. doi:10.1038/nature09710. hdl:10230/25596. PMC 4306417. PMID 21179161.
- Brown, S.; Higham, T.; Slon, V.; Pääbo, S. (2016). "Identification of a new hominin bone from Denisova Cave, Siberia using collagen fingerprinting and mitochondrial DNA analysis". Scientific Reports. 6: 23559. Bibcode:2016NatSR...623559B. doi:10.1038/srep23559. PMC 4810434. PMID 27020421.
- Viola, B. T.; Gunz, P.; Neubauer, S. (2019). "A parietal fragment from Denisova cave". 88th Annual Meeting of the American Association of Physical Anthropologists.
- Lao, O.; Bertranpetit, J.; Mondal, M. (2019). "Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania". Nature Communications. 10 (1): 246. Bibcode:2019NatCo..10..246M. doi:10.1038/s41467-018-08089-7. ISSN 2041-1723. PMC 6335398. PMID 30651539.
- Rogers, A. R.; Bohlender, R. J.; Huff, C. D. (2017). "Early history of Neanderthals and Denisovans". Proceedings of the National Academy of Sciences. 114 (37): 9859–9863. doi:10.1073/pnas.1706426114. PMC 5604018. PMID 28784789.
- Ho, K. K. (2016). "Hominin interbreeding and the evolution of human variation". Journal of Biological Research-Thessaloniki. 23. doi:10.1186/s40709-016-0054-7. PMC 4947341. PMID 27429943.
- Malyarchuk, B. A. (2011). "Adaptive evolution of the Homo mitochondrial genome". Molecular Biology. 45 (5): 845–850. doi:10.1134/S0026893311050104. PMID 22393781.
- Pääbo, S.; Kelso, J.; Reich, D.; Slatkin, M.; Viola, B.; Derevianko, A. P.; Shunkov, M. V.; Doronichev, V. B.; Golovanova, L. V. (2014). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature. 505 (7481): 43–49. Bibcode:2014Natur.505...43P. doi:10.1038/nature12886. ISSN 1476-4687. PMC 4031459. PMID 24352235.
- Kuhlwilm, M.; Gronau, I.; Hubisz, M. J.; de Filippo, C.; Prado-Martinez, J.; Kircher, M.; Fu, Q.; Burbano, H. A.; Lalueza-Fox, C. (2016). "Ancient gene flow from early modern humans into Eastern Neanderthals". Nature. 530 (7591): 429–433. Bibcode:2016Natur.530..429K. doi:10.1038/nature16544. ISSN 1476-4687. PMC 4933530. PMID 26886800.
- Posth, C.; Wißing, C.; Kitagawa, K.; Pagani, L.; van Holstein, L.; Racimo, F.; Wehrberger, K.; Conard, N. J.; Kind, C. J. (2017). "Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals". Nature Communications. 8: 16046. Bibcode:2017NatCo...816046P. doi:10.1038/ncomms16046. ISSN 2041-1723. PMC 5500885. PMID 28675384.
- Bertranpetit, J.; Majumder, P. P.; Li, Q.; Laayouni, H.; Comas, D.; Netea, M. G.; Pybus, M.; Dall'Olio, G. M.; Xu, T. (2016). "Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation". Nature Genetics. 48 (9): 1066–1070. doi:10.1038/ng.3621. hdl:10230/34401. ISSN 1546-1718. PMID 27455350.
- Callaway, E. (2013). "Hominin DNA baffles experts". Nature. 504 (7478): 16–17. Bibcode:2013Natur.504...16C. doi:10.1038/504016a. PMID 24305130.
- Tattersall, I. (2015). The Strange Case of the Rickety Cossack and other Cautionary Tales from Human Evolution. Palgrave Macmillan. p. 200. ISBN 978-1-137-27889-0.
- Meyer, M.; Arsuaga, J.-L.; et al. (2016). "Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins". Nature. 531 (7595): 504–507. Bibcode:2016Natur.531..504M. doi:10.1038/nature17405. PMID 26976447.
- Callaway, E. (2011). "First Aboriginal genome sequenced". Nature News. doi:10.1038/news.2011.551.
- Reich, David; Patterson, Nick; Kircher, Martin; Delfin, Frederick; Nandineni, Madhusudan R; Pugach, Irina; Ko, Albert Min-Shan; Ko, Ying-Chin; Jinam, Timothy A; Phipps, Maude E; Saitou, Naruya; Wollstein, Andreas; Kayser, Manfred; Pääbo, Svante; Stoneking, Mark (2011). "Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania". The American Journal of Human Genetics. 89 (4): 516–28. doi:10.1016/j.ajhg.2011.09.005. PMC 3188841. PMID 21944045.
- Skov, L.; Macià, M. C.; Sveinbjörnsson, G.; et al. (2020). "The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes". Nature. doi:10.1038/s41586-020-2225-9.
- Jacobs, G. S.; Hudjashov, G.; Saag, L.; Kusuma, P.; Darusallam, C. C.; Lawson, D. J.; Mondal, M.; Pagani, L.; Ricaut, F.-X.; Stoneking, M.; Metspalu, M. (2019). "Multiple Deeply Divergent Denisovan Ancestries in Papuans". Cell. 177 (4): 1010–1021.e32. doi:10.1016/j.cell.2019.02.035. ISSN 0092-8674. PMID 30981557.
- Callaway, E. (2019). "Siberia's ancient ghost clan starts to surrender its secrets". Nature News. 566 (7745): 444–446. Bibcode:2019Natur.566..444C. doi:10.1038/d41586-019-00672-2. PMID 30814723.
- Gokhman, D.; Lavi, E.; Prüfer, K.; Fraga, M. F.; Riancho, J. A.; Kelso, J; Pääbo, S; Meshorer, E.; Carmel, L. (2014). "Reconstructing the DNA methylation maps of the Neandertal and the Denisovan". Science. 344 (6183): 523–27. Bibcode:2014Sci...344..523G. doi:10.1126/science.1250368. PMID 24786081.
- Gokhman, D.; et al. (2019). "Reconstructing Denisovan Anatomy Using DNA Methylation Maps". Cell. 179 (1): 180–192. doi:10.1016/j.cell.2019.08.035. PMID 31539495.
- Meyer, M.; Kircher, M.; Gansauge, M.-T.; et al. (2012). "A High-Coverage Genome Sequence from an Archaic Denisovan Individual". Science. 338 (6104): 222–226. Bibcode:2012Sci...338..222M. doi:10.1126/science.1224344. PMC 3617501. PMID 22936568.
- Huerta-Sánchez, E.; Jin, X.; et al. (2014). "Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA". Nature. 512 (7513): 194–97. Bibcode:2014Natur.512..194H. doi:10.1038/nature13408. PMC 4134395. PMID 25043035.
- Fernando Racimo; David Gokhman; Matteo Fumagalli; Amy Ko; Torben Hansen; Ida Moltke; Anders Albrechtsen; Liran Carmel; Emilia Huerta-Sánchez; Rasmus Nielsen (2017). "Archaic Adaptive Introgression in TBX15/WARS2". Molecular Biology and Evolution. 34 (3): 509–524. doi:10.1093/molbev/msw283. PMC 5430617. PMID 28007980.
- Akkuratov, Evgeny E; Gelfand, Mikhail S; Khrameeva, Ekaterina E (2018). "Neanderthal and Denisovan ancestry in Papuans: A functional study". Journal of Bioinformatics and Computational Biology. 16 (2): 1840011. doi:10.1142/S0219720018400115. PMID 29739306.
- Dennell, R. (2019). "Dating of hominin discoveries at Denisova". Nature News. 565 (7741): 571–572. Bibcode:2019Natur.565..571D. doi:10.1038/d41586-019-00264-0. PMID 30700881.
- Pennisi, E. (2013). "More Genomes from Denisova Cave Show Mixing of Early Human Groups". Science. 340 (6134): 799. Bibcode:2013Sci...340..799P. doi:10.1126/science.340.6134.799. PMID 23687020.
- Green RE, Krause J, Briggs AW, et al. (May 2010). "A draft sequence of the Neandertal genome" (PDF). Science. 328 (5979): 710–22. Bibcode:2010Sci...328..710G. doi:10.1126/science.1188021. PMC 5100745. PMID 20448178.
- Prüfer, K.; Racimo, Fe.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; et al. (2013). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature. 505 (7481): 43–49. Bibcode:2014Natur.505...43P. doi:10.1038/nature12886. PMC 4031459. PMID 24352235.
- Warren, Matthew (2018). "Mum's a Neanderthal, Dad's a Denisovan: First discovery of an ancient-human hybrid". Nature. 560 (7719): 417–18. Bibcode:2018Natur.560..417W. doi:10.1038/d41586-018-06004-0. PMID 30135540.
- Wolf, A. B.; Akey, J. M. (2018). "Outstanding questions in the study of archaic hominin admixture". PLOS Genetics. 14 (5): e1007349. doi:10.1371/journal.pgen.1007349. PMC 5978786. PMID 29852022.
- Rogers, A. R.; Harris, N. S.; Achenbach, A. A. (2020). "Neanderthal-Denisovan ancestors interbred with a distantly related hominin". Science Advances. 6 (8): eaay5483. doi:10.1126/sciadv.aay5483. PMC 7032934. PMID 32128408.
- Vernot, B.; et al. (2016). "Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals". Science. 352 (6282): 235–239. Bibcode:2016Sci...352..235V. doi:10.1126/science.aad9416. PMC 6743480. PMID 26989198.
- Rasmussen, M.; et al. (2011). "An Aboriginal Australian genome reveals separate human dispersals into Asia". Science. 334 (6052): 94–98. Bibcode:2011Sci...334...94R. doi:10.1126/science.1211177. PMC 3991479. PMID 21940856.
- Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P. H.; De Filippo, C.; et al. (2013). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature. 505 (7481): 43–49. Bibcode:2014Natur.505...43P. doi:10.1038/nature12886. PMC 4031459. PMID 24352235.
- Browning, S. R.; Browning, B. L.; Zhou, Yi.; Tucci, S.; Akey, J. M. (2018). "Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture". Cell. 173 (1): 53–61.e9. doi:10.1016/j.cell.2018.02.031. ISSN 0092-8674. PMC 5866234. PMID 29551270.
- Fu, Q.; Meyer, M.; Gao, X.; et al. (2013). "DNA analysis of an early modern human from Tianyuan Cave, China". Proceedings of the National Academy of Sciences. 110 (6): 2223–2227. Bibcode:2013PNAS..110.2223F. doi:10.1073/pnas.1221359110. PMC 3568306. PMID 23341637.
- Abi-Rached, L.; Jobin, M. J.; Kulkarni, S.; McWhinnie, A.; Dalva, K.; Gragert, L.; Babrzadeh, F.; Gharizadeh, B.; Luo, M.; Plummer, F. A.; Kimani, J.; Carrington, M.; Middleton, D.; Rajalingam, R.; Beksac, M.; Marsh, S. G. E.; Maiers, M.; Guethlein, L. A.; Tavoularis, S.; Little, A.-M.; Green, R. E.; Norman, P. J.; Parham, P. (2011). "The Shaping of Modern Human Immune Systems by Multiregional Admixture with Archaic Humans". Science. 334 (6052): 89–94. Bibcode:2011Sci...334...89A. doi:10.1126/science.1209202. PMC 3677943. PMID 21868630.
- Sankararaman, S.; Mallick, S.; Patterson, N.; Reich, D. (2016). "The combined landscape of Denisovan and Neanderthal ancestry in present-day humans". Current Biology. 26 (9): 1241–1247. doi:10.1016/j.cub.2016.03.037. PMC 4864120. PMID 27032491.