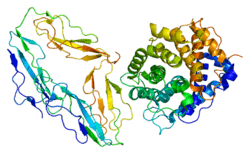
Complement receptor 2
Complement receptor type 2 (CR2), also known as complement C3d receptor, Epstein-Barr virus receptor, and CD21 (cluster of differentiation 21), is a protein that in humans is encoded by the CR2 gene.
CR2 is involved in the complement system. It binds to iC3b (inactive derivative of C3b), C3dg, or C3d.[5] B cells express CR2 receptors on their surfaces, allowing the complement system to play a role in B-cell activation and maturation [6]
Interactions
Complement receptor 2 interacts with CD19,[7][8] and, on mature B cells, forms a complex with CD81 (TAPA-1). The CR2-CD19-CD81 complex is often called the B cell co-receptor complex,[9] because CR2 binds to opsonized antigens through attached C3d (or iC3b or C3dg) when the B-cell receptor binds antigen. This results in the B cell having greatly enhanced response to the antigen.[5]
Epstein-Barr virus (EBV) can bind CR2, enabling EBV to enter and infect B cells. Yefenof et al. (1976) found complete overlapping of EBV receptors and C3 receptors on human B cells.[6][10]
Isoforms
The canonical Cr2/CD21 gene of subprimate mammals produces two types of complement receptor (CR1, ca. 200 kDa; CR2, ca. 145 kDa) via alternative mRNA splicing. The murine Cr2 gene contains 25 exons; a common first exon is spliced to exon 2 and to exon 9 in transcripts encoding CR1 and CR2, respectively. A transcript with an open reading frame of 4,224 nucleotides encodes the long isoform, CR1; this is predicted to be a protein of 1,408 amino acids that includes 21 short consensus repeats (SCR) of ca. 60 amino acids each, plus transmembrane and cytoplasmic regions. Isoform CR2 (1,032 amino acids) is encoded by a shorter transcript (3,096 coding nucleotides) that lacks exons 2-8 encoding SCR1-6. CR1 and CR2 on murine B cells form complexes with a co-accessory activation complex containing CD19, CD81, and the fragilis/Ifitm (murine equivalents of LEU13) proteins.[11]
The CR2 gene of primates produces only the smaller isoform, CR2; primate complement receptor 1, which recapitulates many of the structural domains and presumed functions of Cr2-derived CR1 in subprimates, is encoded by a distinct CR1 gene (apparently derived from the gene Crry of subprimates).
Isoforms CR1 and CR2 derived from the non-primate Cr2 locus possess the same C-terminal sequence, such that association with and activation through CD19 should be equivalent. CR1 can bind to C4b and C3b complexes, whereas CR2 (murine and human) binds to C3dg-bound complexes. CR1, a surface protein produced primarily by follicular dendritic cells, appears to be critical for generation of appropriately activated B cells of the germinal centre and for mature antibody responses to bacterial infection.[12]
Immunohistochemistry
Although CR2 is present on all mature B-cells and follicular dendritic cells (FDCs), this becomes readily apparent only when immunohistochemistry is performed on frozen sections. In more conventional paraffin-embedded tissue samples, only the FDCs retain the staining pattern. As a result, CR2, more commonly called CD21 in the context of immunohistochemistry, can be used to demonstrate the FDC meshwork in lymphoid tissue.
This feature can be useful in examining tissue where the normal germinal centres have been effaced by disease processes, such as HIV infection. The pattern of the FDC meshwork may also be altered in some neoplastic conditions, such as B-cell MALT lymphomas, mantle cell lymphoma, and some T cell lymphomas. Castleman's disease is typified by the presence of abnormal FDCs, and both this, and malignant FDC tumours may therefore be demonstrated using CR2/CD21 antibodies.[13]
References
- GRCh38: Ensembl release 89: ENSG00000117322 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000026616 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Frank K, Atkinson JP (2001). "Complement system." In Austen KF, Frank K, Atkinson JP, Cantor H. eds. Samter's Immunologic Diseases, 6th ed. Vol. 1, p. 281-298, Philadelphia: Lippincott Williams & Wilkins, ISBN 0-7817-2120-2.
- "Entrez Gene: CR2 complement component (3d/Epstein Barr virus) receptor 2".
- Bradbury LE, Kansas GS, Levy S, Evans RL, Tedder TF (November 1992). "The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules". J. Immunol. 149 (9): 2841–50. PMID 1383329.
- Horváth G, Serru V, Clay D, Billard M, Boucheix C, Rubinstein E (November 1998). "CD19 is linked to the integrin-associated tetraspans CD9, CD81, and CD82". J. Biol. Chem. 273 (46): 30537–43. doi:10.1074/jbc.273.46.30537. PMID 9804823.
- Abbas AK, Lichtman AH (2003). Cellular and Molecular Immunology, 5th ed. Philadelphia: Saunders, ISBN 0-7216-0008-5
- Yefenof E, Klein G, Jondal M, Oldstone MB (June 1976). "Surface markers on human B and T-lymphocytes. IX. Two-color immunofluorescence studies on the association between ebv receptors and complement receptors on the surface of lymphoid cell lines". Int. J. Cancer. 17 (6): 693–700. doi:10.1002/ijc.2910170602. PMID 181330.
- Jacobson AC, Weis JH (September 2008). "Comparative functional evolution of human and mouse CR1 and CR2". J. Immunol. 181 (5): 2953–9. doi:10.4049/jimmunol.181.5.2953. PMC 3366432. PMID 18713965.
- Donius LR, Handy JM, Weis JJ, Weis JH (July 2013). "Optimal germinal center B cell activation and T-dependent antibody responses require expression of the mouse complement receptor Cr1". J. Immunol. 191 (1): 434–47. doi:10.4049/jimmunol.1203176. PMC 3707406. PMID 23733878.
- Leong, Anthony S-Y; Cooper, Kumarason; Leong, F Joel W-M (2003). Manual of Diagnostic Cytology (2 ed.). Greenwich Medical Media, Ltd. pp. 93–94. ISBN 978-1-84110-100-2.
Further reading
- Cooper NR, Bradt BM, Rhim JS, Nemerow GR (1990). "CR2 complement receptor". J. Invest. Dermatol. 94 (6 Suppl): s112–s117. doi:10.1111/1523-1747.ep12876069. PMID 2161885.
- Gauffre A, Viron A, Barel M, et al. (1992). "Nuclear localization of the Epstein-Barr virus/C3d receptor (CR2) in the human Burkitt B lymphoma cell, Raji". Mol. Immunol. 29 (9): 1113–20. doi:10.1016/0161-5890(92)90044-X. PMID 1323059.
- Levy E, Ambrus J, Kahl L, et al. (1992). "T lymphocyte expression of complement receptor 2 (CR2/CD21): a role in adhesive cell-cell interactions and dysregulation in a patient with systemic lupus erythematosus (SLE)". Clin. Exp. Immunol. 90 (2): 235–44. doi:10.1111/j.1365-2249.1992.tb07935.x. PMC 1554594. PMID 1424280.
- Matsumoto AK, Kopicky-Burd J, Carter RH, et al. (1991). "Intersection of the complement and immune systems: a signal transduction complex of the B lymphocyte-containing complement receptor type 2 and CD19". J. Exp. Med. 173 (1): 55–64. doi:10.1084/jem.173.1.55. PMC 2118751. PMID 1702139.
- Tuveson DA, Ahearn JM, Matsumoto AK, Fearon DT (1991). "Molecular interactions of complement receptors on B lymphocytes: a CR1/CR2 complex distinct from the CR2/CD19 complex". J. Exp. Med. 173 (5): 1083–9. doi:10.1084/jem.173.5.1083. PMC 2118840. PMID 1708808.
- Kalli KR, Ahearn JM, Fearon DT (1991). "Interaction of iC3b with recombinant isotypic and chimeric forms of CR2". J. Immunol. 147 (2): 590–4. PMID 1830068.
- Barel M, Gauffre A, Lyamani F, et al. (1991). "Intracellular interaction of EBV/C3d receptor (CR2) with p68, a calcium-binding protein present in normal but not in transformed B lymphocytes". J. Immunol. 147 (4): 1286–91. PMID 1831222.
- Delcayre AX, Salas F, Mathur S, et al. (1991). "Epstein Barr virus/complement C3d receptor is an interferon alpha receptor". EMBO J. 10 (4): 919–26. doi:10.1002/j.1460-2075.1991.tb08025.x. PMC 452735. PMID 1849076.
- Kurtz CB, Paul MS, Aegerter M, et al. (1989). "Murine complement receptor gene family. II. Identification and characterization of the murine homolog (Cr2) to human CR2 and its molecular linkage to Crry". J. Immunol. 143 (6): 2058–67. PMID 2528587.
- Fujisaku A, Harley JB, Frank MB, et al. (1989). "Genomic organization and polymorphisms of the human C3d/Epstein-Barr virus receptor". J. Biol. Chem. 264 (4): 2118–25. PMID 2563370.
- Moore MD, Cooper NR, Tack BF, Nemerow GR (1988). "Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes". Proc. Natl. Acad. Sci. U.S.A. 84 (24): 9194–8. doi:10.1073/pnas.84.24.9194. PMC 299719. PMID 2827171.
- Weis JJ, Toothaker LE, Smith JA, et al. (1988). "Structure of the human B lymphocyte receptor for C3d and the Epstein-Barr virus and relatedness to other members of the family of C3/C4 binding proteins". J. Exp. Med. 167 (3): 1047–66. doi:10.1084/jem.167.3.1047. PMC 2188894. PMID 2832506.
- Weis JJ, Fearon DT, Klickstein LB, et al. (1986). "Identification of a partial cDNA clone for the C3d/Epstein-Barr virus receptor of human B lymphocytes: homology with the receptor for fragments C3b and C4b of the third and fourth components of complement". Proc. Natl. Acad. Sci. U.S.A. 83 (15): 5639–43. doi:10.1073/pnas.83.15.5639. PMC 386344. PMID 3016712.
- Weis JH, Morton CC, Bruns GA, et al. (1987). "A complement receptor locus: genes encoding C3b/C4b receptor and C3d/Epstein-Barr virus receptor map to 1q32". J. Immunol. 138 (1): 312–5. PMID 3782802.
- Aubry JP, Pochon S, Gauchat JF, et al. (1994). "CD23 interacts with a new functional extracytoplasmic domain involving N-linked oligosaccharides on CD21". J. Immunol. 152 (12): 5806–13. PMID 7515913.
- Matsumoto AK, Martin DR, Carter RH, et al. (1993). "Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes". J. Exp. Med. 178 (4): 1407–17. doi:10.1084/jem.178.4.1407. PMC 2191213. PMID 7690834.
- Barel M, Balbo M, Gauffre A, Frade R (1995). "Binding sites of the Epstein-Barr virus and C3d receptor (CR2, CD21) for its three intracellular ligands, the p53 anti-oncoprotein, the p68 calcium binding protein and the nuclear p120 ribonucleoprotein". Mol. Immunol. 32 (6): 389–97. doi:10.1016/0161-5890(95)00005-Y. PMID 7753047.
External links
- Complement+Receptors+2 at the US National Library of Medicine Medical Subject Headings (MeSH)