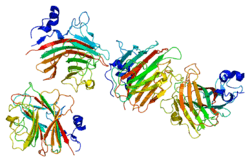LMAN1
Protein ERGIC-53 also known as ER-Golgi intermediate compartment 53 kDa protein or lectin mannose-binding 1 is a protein that in humans is encoded by the LMAN1 gene.[5][6][7]
Function
ERGIC-53 (also named LMAN1) is a type I integral membrane protein localized in the intermediate region (ERGIC) between the endoplasmic reticulum and the Golgi, presumably recycling between the two compartments. The protein is a mannose-specific lectin and is a member of a novel family of plant lectin homologs in the secretory pathway of animal cells. Mutations in the gene are associated with a coagulation defect. Using positional cloning, the gene was identified as the disease gene leading to combined deficiency of factor V-factor VIII, a rare, autosomal recessive disorder in which both coagulation factors V and VIII are diminished.[8][7] MCFD2 is the second gene that leads to combined deficiency of factor V-factor VIII.[9] ERGIC-53 and MCFD2 form a protein complex and serve as a cargo receptor to transport FV and FVIII from the ER to the ERGIC and then the Golgi,[10]as illustrated here.[8]
Clinical significance
LMAN1 mutational inactivation is a frequent and early event potentially contributing to colorectal tumorigenesis.[11]
References
- GRCh38: Ensembl release 89: ENSG00000074695 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000041891 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, Moussalli MJ, Hauri HP, Ciavarella N, Kaufman RJ, Ginsburg D (May 1998). "Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII". Cell. 93 (1): 61–70. doi:10.1016/S0092-8674(00)81146-0. PMID 9546392.
- Arar C, Mignon C, Mattei M, Monsigny M, Roche A, Legrand A (Feb 1997). "Mapping of the MR60/ERGIC-53 gene to human chromosome 18q21.3-18q22 by in situ hybridization". Mamm Genome. 7 (10): 791–2. doi:10.1007/s003359900238. PMID 8854877.
- "Entrez Gene: LMAN1 lectin, mannose-binding, 1".
- Khoriaty R, Vasievich MP, Ginsburg D (July 2012). "The COPII pathway and hematologic disease". Blood. 120 (1): 31–8. doi:10.1182/blood-2012-01-292086. PMC 3390960. PMID 22586181.
- Zhang B, Cunningham MA, Nichols WC, Bernat JA, Seligsohn U, Pipe SW, McVey JH, Schulte-Overberg U, de Bosch NB, Ruiz-Saez A, White GC, Tuddenham EG, Kaufman RJ, Ginsburg D (May 2003). "Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex". Nat Genet. 34 (2): 220–5. doi:10.1038/ng1153. PMID 12717434.
- Zhang B, Kaufman RJ, Ginsburg D (2005). "LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway". J. Biol. Chem. 280 (27): 25881–6. doi:10.1074/jbc.M502160200. PMID 15886209.
- Roeckel N, Woerner SM, Kloor M, Yuan YP, Patsos G, Gromes R, Kopitz J, Gebert J (January 2009). "High frequency of LMAN1 abnormalities in colorectal tumors with microsatellite instability". Cancer Res. 69 (1): 292–9. doi:10.1158/0008-5472.CAN-08-3314. PMID 19118014.
Further reading
- Botos I, Wlodawer A (2005). "Proteins that bind high-mannose sugars of the HIV envelope". Prog. Biophys. Mol. Biol. 88 (2): 233–82. doi:10.1016/j.pbiomolbio.2004.05.001. PMID 15572157.
- Arar C, Carpentier V, Le Caer JP, et al. (1995). "ERGIC-53, a membrane protein of the endoplasmic reticulum-Golgi intermediate compartment, is identical to MR60, an intracellular mannose-specific lectin of myelomonocytic cells". J. Biol. Chem. 270 (8): 3551–3. doi:10.1074/jbc.270.8.3551. PMID 7876089.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Fiedler K, Simons K (1994). "A putative novel class of animal lectins in the secretory pathway homologous to leguminous lectins". Cell. 77 (5): 625–6. doi:10.1016/0092-8674(94)90047-7. PMID 8205612.
- Schindler R, Itin C, Zerial M, et al. (1993). "ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif". Eur. J. Cell Biol. 61 (1): 1–9. PMID 8223692.
- Haurum JS, Thiel S, Jones IM, et al. (1994). "Complement activation upon binding of mannan-binding protein to HIV envelope glycoproteins". AIDS. 7 (10): 1307–13. doi:10.1097/00002030-199310000-00002. PMID 8267903.
- Senaldi G, Davies ET, Mahalingam M, et al. (1996). "Circulating levels of mannose binding protein in human immunodeficiency virus infection". J. Infect. 31 (2): 145–8. doi:10.1016/S0163-4453(95)92185-0. PMID 8666845.
- Neerman-Arbez M, Antonarakis SE, Blouin JL, et al. (1997). "The locus for combined factor V-factor VIII deficiency (F5F8D) maps to 18q21, between D18S849 and D18S1103". Am. J. Hum. Genet. 61 (1): 143–50. doi:10.1086/513897. PMC 1715850. PMID 9245995.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, et al. (1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Nichols WC, Terry VH, Wheatley MA, et al. (1999). "ERGIC-53 gene structure and mutation analysis in 19 combined factors V and VIII deficiency families". Blood. 93 (7): 2261–6. PMID 10090935.
- Saifuddin M, Hart ML, Gewurz H, et al. (2000). "Interaction of mannose-binding lectin with primary isolates of human immunodeficiency virus type 1". J. Gen. Virol. 81 (Pt 4): 949–55. doi:10.1099/0022-1317-81-4-949. PMID 10725420.
- Harris RA, Yang A, Stein RC, et al. (2002). "Cluster analysis of an extensive human breast cancer cell line protein expression map database". Proteomics. 2 (2): 212–23. doi:10.1002/1615-9861(200202)2:2<212::AID-PROT212>3.0.CO;2-H. PMID 11840567.
- Jüliger S, Kremsner PG, Alpers MP, et al. (2002). "Restricted polymorphisms of the mannose-binding lectin gene in a population of Papua New Guinea". Mutat. Res. 505 (1–2): 87–91. doi:10.1016/S0027-5107(02)00142-2. PMID 12175909.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Hart ML, Saifuddin M, Uemura K, et al. (2003). "High mannose glycans and sialic acid on gp120 regulate binding of mannose-binding lectin (MBL) to HIV type 1". AIDS Res. Hum. Retroviruses. 18 (17): 1311–7. doi:10.1089/088922202320886352. PMID 12487819.
- Hart ML, Saifuddin M, Spear GT (2003). "Glycosylation inhibitors and neuraminidase enhance human immunodeficiency virus type 1 binding and neutralization by mannose-binding lectin". J. Gen. Virol. 84 (Pt 2): 353–60. doi:10.1099/vir.0.18734-0. PMID 12560567.
- Gevaert K, Goethals M, Martens L, et al. (2004). "Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides". Nat. Biotechnol. 21 (5): 566–9. doi:10.1038/nbt810. PMID 12665801.
- García-Laorden MI, Rúa-Figueroa I, Pérez-Aciego P, et al. (2003). "Mannose binding lectin polymorphisms as a disease-modulating factor in women with systemic lupus erythematosus from Canary Islands, Spain". J. Rheumatol. 30 (4): 740–6. PMID 12672193.