Plasma membrane H+-ATPase
The P-type plasma membrane H+
-ATPase is found in plants and fungi. For the gastric H+
/K+
ATPase (involved in the acidification of the stomach in mammals), see Hydrogen potassium ATPase.
| hydrogen-exporting ATPase, phosphorylative mechanism | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.6.3.6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Identifiers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | E1-E2_ATPase | ||||||||||
| Pfam | PF00122 | ||||||||||
| InterPro | IPR000695 | ||||||||||
| PROSITE | PDOC00139 | ||||||||||
| TCDB | 3.A.3.3 | ||||||||||
| OPM superfamily | 22 | ||||||||||
| OPM protein | 4hqj | ||||||||||
| |||||||||||
Plasma membrane H+
-ATPase (P-type)
-ATPase (P-type)
This enzyme belongs to the family of hydrolases, specifically those acting on acid anhydrides to catalyse transmembrane movement of substances. To be specific, the protein is a part of the P-type ATPase family. The systematic name of this enzyme class is ATP phosphohydrolase (H+
-exporting).
H+
-exporting ATPase is also known as proton ATPase or more simply proton pump. Other names in common use include proton-translocating ATPase, yeast plasma membrane H+
-ATPase, plant plasma membrane H+
-ATPase, yeast plasma membrane ATPase, plant plasma membrane ATPase, and ATP phosphohydrolase.
The yeast (Saccharomyces cerevisiae) enzyme is encoded by the gene Pma1 and hence referred to as Pma1p.[1]
Function and location
The plasma membrane H+
-ATPase or proton pump creates the electrochemical gradients in the plasma membrane of plants, fungi, protists, and many prokaryotes. Here, proton gradients are used to drive secondary transport processes. As such, it is essential for the uptake of most metabolites, and also for plant responses to the environment (e.g., movement of leaves).
H+
-ATPases are specific for plants, fungi, and protists; and Na+
/K+
-ATPases are specific for animal cells. These two groups of P-type ATPases, although not from the same subfamily, seem to perform a complementary function in plants/fungi/protists and animal cells, namely the creation of an electrochemical gradient used as an energy source for secondary transport.[2]
Structural studies
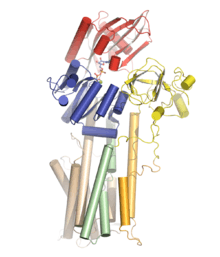
Structural information on P-type plasma membrane (PM) proton ATPases are scarce compared to that obtained for SERCA1a. A low resolution structure from 2D crystals of the PM H+
-ATPase from Neurospora crassa is, as of medio 2011, the only structural information on the fungal H+
-ATPase.[3] For the plant counterpart, a crystal structure of the AHA2 PM H+
-ATPase from Arabidopsis thaliana has been obtained from 3D crystals with a resolution of 3.6 Å.[4] The structure of AHA2 clearly identifies three cytosolic domains corresponding to the N (nucleotide binding), P (phosphorylation), and A (actuator) domains, similar to those observed in the SR Ca2+
-ATPase and also verifies the presence of ten transmembrane helices. The 3D crystal structure shows the AHA2 PM H+
-ATPase in a so-called quasi-occluded E1 state with the non-hydrolysable ATP analogue AMPPCP bound, and the overall fold of the catalytic unit reveals a high degree of structural similarity to the SR Ca2+
-ATPase and the Na+
,K+
-ATPase. The overall arrangement of the domains is similar to that observed for the occluded E1 conformation of the SR Ca2+
-ATPase, and based on comparison with structural data for the other conformations of the SR Ca2+
-ATPase, it was suggested that the structure of the AHA2 PM H+
-ATPase represents a novel E1 intermediate.[4] A distinct feature of the PM H+
-ATPase not observed in other P-type ATPases is the presence of a large cavity in the transmembrane domain formed by M4, M5 and M6.
Regulation
Precise regulation of PM H+
-ATPase activity is crucial to the plant. Over-expression of the PM H+
-ATPase is compensated by a down-regulation of activity,[5] whereas deletion of an isoform is compensated by redundancy as well as augmented activity of other isoforms by increased level of post-translational modifications.[6]
The PM H+
-ATPase is subject to autoinhibition, which negatively regulates the activity of the pump and keeps the enzyme in a low activity state where ATP hydrolytic activity is partly uncoupled from ATP hydrolysis,.[7][8] Release from the autoinhibitory restraints requires posttranslational modifications such as phosphorylation and interacting proteins.
Autoinhibition is achieved by the N- and C-termini of the protein - communication between the two termini facilitates the necessary precise control of pump activity.[9] The autoinhibitory C-terminal domain can be displaced by phosphorylation of the penultimate Thr residue and the subsequent binding of 14-3-3 proteins.[10][11] The PM H+
-ATPase is the first P-type ATPase for which both termini have been demonstrated to take part in the regulation of protein activity.[9]
Physiological roles in plants
Plasma membrane H+
-ATPases are found throughout the plant in all cell types investigated, but some cell types have much higher concentrations of H+
-ATPase than others. In general, these cell types are specialised for intensive active transport and accumulate solutes from their surroundings. Most studies of these roles come from genetic studies on Arabidopsis thaliana.[12] H+
-ATPases in plants are expressed from a multigene subfamily, and Arabidopsis thaliana for instance, have 12 different H+
-ATPase genes.
Some important physiological processes the plant H+
-ATPase is involved in are:
- Phloem loading. The phloem is a tissue specialised for long-distance transport of organic compounds, and is well known for its involvement in the transport of sugar from leaves or other source areas. Here the H+
-ATPase powers the sucrose/H+
cotransporters and is found to be essential for the loading of sucrose into the phloem. - Solute uptake in roots. H+
-ATPases energize the uptake of nutrients from the soil into the roots, and is also involved in the further loading of these solutes into the xylem, a tissue specialised for long-distance transport of water and micronutrients. - Tip-growing systems. Pollen tubes and root hairs are examples of plant tip-growing systems, where a single cell expands in one direction only. The direction of growth is controlled by an asymmetrical proton gradient, where protons enter at the extreme tip and are pumped out just below the tip.
- Size of stomatal aperture. The somatal pore controls the diffusion of CO
2 into the leaves to be utilized for photosynthesis. The pore is formed by two guard cells, which control the size of the pore by swelling in response to the activity of the H+
-ATPase. Opening and closure of the pore is partly controlled by regulation of the H+
-ATPase. - Plant movements. Like the somatal pore, other movements of plant organs are controlled by motor cells' changing cell turgor. These cells control phenomena such as solar tracking by the plant to optimize orientation of photosynthetic leaves, and the swift and spectacular reactions to touch found in some plant species (e.g., carnivorous plants). All of these swelling and shrinking processes take place by massive water and ion fluxes through channels. Here, activation of the H+
-ATPase leads to plasma membrane hyperpolarization and the opening of voltage sensitive potassium channels. The K+
influx leads to water uptake and turgor increase in the cell. - Salt and osmotolerance. Salinity imposes two stresses on the cell: one is the loss of turgor due to the hypertonicity of the extracellular medium, and the other is a direct effect of toxic ions on metabolism. Therefore, plants have developed several defence mechanisms. The Na/H+
antiporter is heavily involved and is powered by the action of the H+
-ATPase, which is highly expressed in leaves and roots during salt stress. - Intracellular pH regulation. Intracellular pH remains constant during cell growth, it is presumed, to ensure optimal activity of the cytoplasmic enzymes. This is controlled by the proton pump.
- Acid growth. Acidification of the external medium caused by activation of the plasma membrane H+
-ATPase initiates cellular expansion. It is believed that the plant hormone auxin activates the proton pump. The apoplastic acidification leads to loosening of the cell wall and hyperpolarization of the plasma membrane inducing K+
uptake and swelling.
References
- Thierry Ferreira, A. Brett Mason and Carolyn W. Slayman (2001). "The Yeast Pma1 Proton Pump: a Model for Understanding the Biogenesis of Plasma Membrane Proteins". J Biol Chem. 276 (32): 29613–29616. doi:10.1074/jbc.R100022200. PMID 11404364.
- Pedersen JT, Palmgren M (March 2017). "Why do plants lack sodium pumps and would they benefit from having one?". Functional Plant Biology. 44 (5): 473–479. doi:10.1071/FP16422.
- Auer M, Scarborough GA, Kühlbrandt W (April 1998). "Three-dimensional map of the plasma membrane H+
-ATPase in the open conformation". Nature. 392 (6678): 840–3. doi:10.1038/33967. PMID 9572146. - Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P (December 2007). "Crystal structure of the plasma membrane proton pump". Nature. 450 (7172): 1111–4. Bibcode:2007Natur.450.1111P. doi:10.1038/nature06417. PMID 18075595.
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M (August 2007). "Expression of a constitutively activated plasma membrane H+
-ATPase alters plant development and increases salt tolerance". Plant Physiol. 144 (4): 1763–76. doi:10.1104/pp.107.103762. PMC 1949876. PMID 17600134. - Haruta M, Burch HL, Nelson RB, et al. (June 2010). "Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity". J. Biol. Chem. 285 (23): 17918–29. doi:10.1074/jbc.M110.101733. PMC 2878554. PMID 20348108.
- Palmgren MG, Sommarin M, Serrano R, Larsson C (October 1991). "Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+
-ATPase". J. Biol. Chem. 266 (30): 20470–5. PMID 1834646. - Morsomme P, de Kerchove d'Exaerde A, De Meester S, Thinès D, Goffeau A, Boutry M (October 1996). "Single point mutations in various domains of a plant plasma membrane H+
-ATPase expressed in Saccharomyces cerevisiae increase H+
-pumping and permit yeast growth at low pH". EMBO J. 15 (20): 5513–26. doi:10.1002/j.1460-2075.1996.tb00936.x. PMC 452296. PMID 8896445. - Ekberg K, Palmgren MG, Veierskov B, Buch-Pedersen MJ (March 2010). "A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein". J. Biol. Chem. 285 (10): 7344–50. doi:10.1074/jbc.M109.096123. PMC 2844182. PMID 20068040.
- Svennelid F, Olsson A, Piotrowski M, et al. (December 1999). "Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+
-ATPase creates a binding site for the regulatory 14-3-3 protein". Plant Cell. 11 (12): 2379–91. doi:10.2307/3870962. JSTOR 3870962. PMC 144135. PMID 10590165. - Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG (December 1999). "Binding of 14-3-3 protein to the plasma membrane H+
-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947". J Biol Chem. 274 (51): 36774–80. doi:10.1074/jbc.274.51.36774. PMID 10593986. - Palmgren MG (June 2001). "PLANT PLASMA MEMBRANE H+
-ATPases: Powerhouses for Nutrient Uptake". Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 817–845. doi:10.1146/annurev.arplant.52.1.817. PMID 11337417.
- Goffeau A, Slayman CW (1981). "The proton-translocating ATPase of the fungal plasma membrane". Biochim. Biophys. Acta. 639 (3–4): 197–223. doi:10.1016/0304-4173(81)90010-0. PMID 6461354.
- Serrano R, Kielland-Brandt MC, Fink GR (1986). "Yeast plasma membrane ATPase is essential for growth and has homology with (Na+
+ K+
), K+
- and Ca2+
-ATPases". Nature. 319 (6055): 689–93. doi:10.1038/319689a0. PMID 3005867. - Serrano R, Portillo F (1990). "Catalytic and regulatory sites of yeast plasma membrane H+
-ATPase studied by directed mutagenesis". Biochim. Biophys. Acta. 1018 (2–3): 195–9. doi:10.1016/0005-2728(90)90247-2. PMID 2144186.