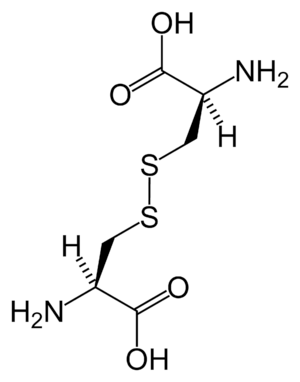
Disulfide
In chemistry, a disulfide refers to a functional group with the structure R−S−S−R′. The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In biology, disulfide bridges formed between thiol groups in two cysteine residues are an important component of the secondary and tertiary structure of proteins. The connection is a persulfide, in analogy to its congener, peroxide (R−O−O−R′), but this terminology is rarely used, except in reference to hydrodisulfides (R−S−S−H compounds).
In inorganic chemistry disulfide usually refers to the corresponding anion S2−
2 (−S−S−).
Organic disulfides

Symmetrical disulfides are compounds of the formula R2S2. Most disulfides encountered in organo sulfur chemistry are symmetrical disulfides. Unsymmetrical disulfides (also called heterodisulfides) are compounds of the formula RSSR'. They are less common in organic chemistry, but most disulfides in nature are unsymmetrical.
Properties
The disulfide bonds are strong, with a typical bond dissociation energy of 60 kcal/mol (251 kJ mol−1). However, being about 40% weaker than C−C and C−H bonds, the disulfide bond is often the "weak link" in many molecules. Furthermore, reflecting the polarizability of divalent sulfur, the S−S bond is susceptible to scission by polar reagents, both electrophiles and especially nucleophiles (Nu):[1]
- RS−SR + Nu− → RS−Nu + RS−
The disulfide bond is about 2.05 Å in length, about 0.5 Å longer than a C−C bond. Rotation about the S−S axis is subject to a low barrier. Disulfides show a distinct preference for dihedral angles approaching 90°. When the angle approaches 0° or 180°, then the disulfide is a significantly better oxidant.
Disulfides where the two R groups are the same are called symmetric, examples being diphenyl disulfide and dimethyl disulfide. When the two R groups are not identical, the compound is said to be an asymmetric or mixed disulfide.[2]
Although the hydrogenation of disulfides is usually not practical, the equilibrium constant for the reaction provides a measure of the standard redox potential for disulfides:
- RSSR + H2 → 2 RSH
This value is about −250 mV versus the standard hydrogen electrode (pH = 7). By comparison, the standard reduction potential for ferrodoxins is about −430 mV.
Synthesis
Disulfide bonds are usually formed from the oxidation of sulfhydryl (−SH) groups, especially in biological contexts.[3] The transformation is depicted as follows:
- 2 RSH ⇌ RS−SR + 2 H+ + 2 e−
A variety of oxidants participate in this reaction including oxygen and hydrogen peroxide. Such reactions are thought to proceed via sulfenic acid intermediates. In the laboratory, iodine in the presence of base is commonly employed to oxidize thiols to disulfides. Several metals, such as copper(II) and iron(III) complexes affect this reaction. Alternatively, disulfide bonds in proteins often formed by thiol-disulfide exchange:
- RS−SR + R′SH ⇌ R′S−SR + RSH
Such reactions are mediated by enzymes in some cases and in other cases are under equilibrium control, especially in the presence of a catalytic amount of base.
The alkylation of alkali metal di- and polysulfides gives disulfides. "Thiokol" polymers arise when sodium polysulfide is treated with an alkyl dihalide. In the converse reaction, carbanionic reagents react with elemental sulfur to afford mixtures of the thioether, disulfide, and higher polysulfides. These reactions are often unselective but can be optimized for specific applications.
Synthesis of unsymmetrical disulfides (heterodisulfides)
Many specialized methods have been developed for forming unsymmetrical disulfides. Reagents that deliver the equivalent of "RS+" react with thiols to give asymmetrical disulfides:[3]
- RSH + R′SNR″2 → RS−SR′ + HNR″2
where R″2N is the phthalimido group. Bunte salts, derivatives of the type RSSO3–Na+ are also used to generate unsymmetrical disulfides:[4] Na[O3S2R] + NaSR' → RSSR' + Na2SO3
Reactions
The most important aspect of disulfide bonds is their cleavage, which occurs via reduction. A variety of reductants can be used. In biochemistry, thiols such as β-mercaptoethanol (β-ME) or dithiothreitol (DTT) serve as reductants; the thiol reagents are used in excess to drive the equilibrium to the right:
- RS−SR + 2 HOCH2CH2SH ⇌ HOCH2CH2S−SCH2CH2OH + 2 RSH
The reductant tris(2-carboxyethyl)phosphine (TCEP) is useful, beside being odorless compared to β-ME and DTT, because it is selective, working at both alkaline and acidic conditions (unlike DTT), is more hydrophilic and more resistant to oxidation in air. Furthermore, it is often not needed to remove TCEP before modification of protein thiols.[5]
In organic synthesis, hydride agents are typically employed for scission of disulfides, such as sodium borohydride. More aggressive, alkali metals will effect this reaction:
- RS−SR + 2 Na → 2 NaSR
These reactions are often followed by protonation of the resulting metal thiolate:
- NaSR + HCl → HSR + NaCl
Thiol–disulfide exchange is a chemical reaction in which a thiolate group −S− attacks a sulfur atom of a disulfide bond −S−S−. The original disulfide bond is broken, and its other sulfur atom is released as a new thiolate, carrying away the negative charge. Meanwhile, a new disulfide bond forms between the attacking thiolate and the original sulfur atom.[6][7]
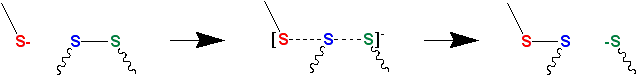
Thiolates, not thiols, attack disulfide bonds. Hence, thiol–disulfide exchange is inhibited at low pH (typically, below 8) where the protonated thiol form is favored relative to the deprotonated thiolate form. (The pKa of a typical thiol group is roughly 8.3, but can vary due to its environment.)
Thiol–disulfide exchange is the principal reaction by which disulfide bonds are formed and rearranged in a protein. The rearrangement of disulfide bonds within a protein generally occurs via intra-protein thiol–disulfide exchange reactions; a thiolate group of a cysteine residue attacks one of the protein's own disulfide bonds. This process of disulfide rearrangement (known as disulfide shuffling) does not change the number of disulfide bonds within a protein, merely their location (i.e., which cysteines are bonded). Disulfide reshuffling is generally much faster than oxidation/reduction reactions, which change the number of disulfide bonds within a protein. The oxidation and reduction of protein disulfide bonds in vitro also generally occurs via thiol–disulfide exchange reactions. Typically, the thiolate of a redox reagent such as glutathione or dithiothreitol attacks the disulfide bond on a protein forming a mixed disulfide bond between the protein and the reagent. This mixed disulfide bond when attacked by another thiolate from the reagent, leaves the cysteine oxidized. In effect, the disulfide bond is transferred from the protein to the reagent in two steps, both thiol–disulfide exchange reactions.
The in vivo oxidation and reduction of protein disulfide bonds by thiol–disulfide exchange is facilitated by a protein called thioredoxin. This small protein, essential in all known organisms, contains two cysteine amino acid residues in a vicinal arrangement (i.e., next to each other), which allows it to form an internal disulfide bond, or disulfide bonds with other proteins. As such, it can be used as a repository of reduced or oxidized disulfide bond moieties.
Many specialized organic reactions have been developed for disulfides, again mainly associated with the scission of the S−S bond, which is usually the weakest bond in a molecule. In the Zincke disulfide cleavage reactions, disulfides are cleaved by halogens. This reaction gives a sulfenyl halide:[8][9]
- ArSSAr + Cl2 → 2 ArSCl
Occurrence in biology
_V.1.svg.png)
Occurrence in proteins
Disulfide bonds can be formed under oxidising conditions and play an important role in the folding and stability of some proteins, usually proteins secreted to the extracellular medium.[2] Since most cellular compartments are reducing environments, in general, disulfide bonds are unstable in the cytosol, with some exceptions as noted below, unless a sulfhydryl oxidase is present.[10]
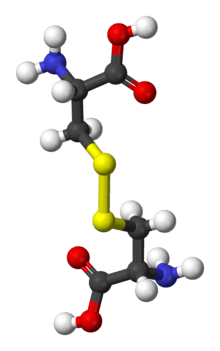
Disulfide bonds in proteins are formed between the thiol groups of cysteine residues by the process of oxidative folding. The other sulfur-containing amino acid, methionine, cannot form disulfide bonds. A disulfide bond is typically denoted by hyphenating the abbreviations for cysteine, e.g., when referring to ribonuclease A the "Cys26–Cys84 disulfide bond", or the "26–84 disulfide bond", or most simply as "C26–C84"[11] where the disulfide bond is understood and does not need to be mentioned. The prototype of a protein disulfide bond is the two-amino-acid peptide cystine, which is composed of two cysteine amino acids joined by a disulfide bond. The structure of a disulfide bond can be described by its χss dihedral angle between the Cβ−Sγ−Sγ−Cβ atoms, which is usually close to ±90°.
The disulfide bond stabilizes the folded form of a protein in several ways:
- It holds two portions of the protein together, biasing the protein towards the folded topology. That is, the disulfide bond destabilizes the unfolded form of the protein by lowering its entropy.
- The disulfide bond may form the nucleus of a hydrophobic core of the folded protein, i.e., local hydrophobic residues may condense around the disulfide bond and onto each other through hydrophobic interactions.
- Related to 1 and 2, the disulfide bond link two segments of the protein chain, the disulfide bond increases the effective local concentration of protein residues and lowers the effective local concentration of water molecules. Since water molecules attack amide-amide hydrogen bonds and break up secondary structure, a disulfide bond stabilizes secondary structure in its vicinity. For example, researchers have identified several pairs of peptides that are unstructured in isolation, but adopt stable secondary and tertiary structure upon forming a disulfide bond between them.
A disulfide species is a particular pairing of cysteines in a disulfide-bonded protein and is usually depicted by listing the disulfide bonds in parentheses, e.g., the "(26–84, 58–110) disulfide species". A disulfide ensemble is a grouping of all disulfide species with the same number of disulfide bonds, and is usually denoted as the 1S ensemble, the 2S ensemble, etc. for disulfide species having one, two, etc. disulfide bonds. Thus, the (26–84) disulfide species belongs to the 1S ensemble, whereas the (26–84, 58–110) species belongs to the 2S ensemble. The single species with no disulfide bonds is usually denoted as R for "fully reduced". Under typical conditions, disulfide reshuffling is much faster than the formation of new disulfide bonds or their reduction; hence, the disulfide species within an ensemble equilibrate more quickly than between ensembles.
The native form of a protein is usually a single disulfide species, although some proteins may cycle between a few disulfide states as part of their function, e.g., thioredoxin. In proteins with more than two cysteines, non-native disulfide species may be formed, which are almost always misfolded. As the number of cysteines increases, the number of nonnative species increases factorially.
In bacteria and archaea
Disulfide bonds play an important protective role for bacteria as a reversible switch that turns a protein on or off when bacterial cells are exposed to oxidation reactions. Hydrogen peroxide (H2O2) in particular could severely damage DNA and kill the bacterium at low concentrations if not for the protective action of the SS-bond. Archaea typically have fewer disulfides than higher organisms.[12]
In eukaryotes
In eukaryotic cells, in general, stable disulfide bonds are formed in the lumen of the RER (rough endoplasmic reticulum) and the mitochondrial intermembrane space but not in the cytosol. This is due to the more oxidizing environment of the aforementioned compartments and more reducing environment of the cytosol (see glutathione). Thus disulfide bonds are mostly found in secretory proteins, lysosomal proteins, and the exoplasmic domains of membrane proteins.
There are notable exceptions to this rule. For example, many nuclear and cytosolic proteins can become disulfide-crosslinked during necrotic cell death.[13] Similarly, a number of cytosolic proteins which have cysteine residues in proximity to each other that function as oxidation sensors or redox catalysts; when the reductive potential of the cell fails, they oxidize and trigger cellular response mechanisms. The virus Vaccinia also produces cytosolic proteins and peptides that have many disulfide bonds; although the reason for this is unknown presumably they have protective effects against intracellular proteolysis machinery.
Disulfide bonds are also formed within and between protamines in the sperm chromatin of many mammalian species.
Disulfides in regulatory proteins
As disulfide bonds can be reversibly reduced and re-oxidized, the redox state of these bonds has evolved into a signaling element. In chloroplasts, for example, the enzymatic reduction of disulfide bonds has been linked to the control of numerous metabolic pathways as well as gene expression. The reductive signaling activity has been shown, thus far, to be carried by the ferredoxin thioredoxin system, channeling electrons from the light reactions of photosystem I to catalytically reduce disulfides in regulated proteins in a light dependent manner. In this way chloroplasts adjust the activity of key processes such as the Calvin–Benson cycle, starch degradation, ATP production and gene expression according to light intensity.
In hair and feathers
Over 90% of the dry weight of hair comprises proteins called keratins, which have a high disulfide content, from the amino acid cysteine. The robustness conferred in part by disulfide linkages is illustrated by the recovery of virtually intact hair from ancient Egyptian tombs. Feathers have similar keratins and are extremely resistant to protein digestive enzymes. The stiffness of hair and feather is determined by the disulfide content. Manipulating disulfide bonds in hair is the basis for the permanent wave in hairstyling. Reagents that affect the making and breaking of S−S bonds are key, e.g., ammonium thioglycolate. The high disulfide content of feathers dictates the high sulfur content of bird eggs. The high sulfur content of hair and feathers contributes to the disagreeable odor that results when they are burned.
Inorganic disulfides
The disulfide anion is S2−
2, or −S−S−. In disulfide, sulfur exists in the reduced state with oxidation number −1. Its electron configuration then resembles that of a chlorine atom. It thus tends to form a covalent bond with another S− center to form S2−
2 group, similar to elemental chlorine existing as the diatomic Cl2. Oxygen may also behave similarly, e.g. in peroxides such as H2O2. Examples:
- Hydrogen disulfide (S2H2), the simplest inorganic disulfide
- Disulfur dichloride (S2Cl2), a distillable liquid.
- Iron disulfide (FeS2)
In industry
Disulfide and (polysulfide) bonds are the crosslinking groups that result from the vulcanization of rubber. In analogy to the role of disulfides in proteins, the S−S linkages in rubber are crosslinkers, and strongly affect the rheology of the material.
Related compounds


Thiosulfoxides are orthogonally isomeric with disulfides, having the second sulfur branching from the first and not partaking in a continuous chain, i.e. >S=S rather than −S−S−.
Disulfide bonds are analogous but more common than related peroxide, thioselenide, and diselenide bonds. Intermediate compounds of these also exist, for example thioperoxides (also known as oxasulfides) such as hydrogen thioperoxide, have the formula R1OSR2 (equivalently R2SOR1). These are isomeric to sulfoxides in a similar manner to the above; i.e. >S=O rather than −S−O−.
Thiuram disulfides, with the formula (R2NCSS)2, are disulfides but they behave distinctly because of the thiocarbonyl group.
Compounds with three sulfur atoms, such as CH3S−S−SCH3, are called trisulfides, or trisulfide bonds.
Misnomers
Disulfide is also used to refer to compounds that contain two sulfide (S2−) centers. The compound carbon disulfide, CS2 is described with the structural formula i.e. S=C=S. This molecule is not a disulfide in the sense that it lacks a S-S bond. Similarly, molybdenum disulfide, MoS2, is not a disulfide in the sense again that its sulfur atoms are not linked.
References
- Cremlyn, R. J. (1996). An Introduction to Organosulfur Chemistry. Chichester: John Wiley and Sons. ISBN 0-471-95512-4.
- Sevier, C. S.; Kaiser, C. A. (2002). "Formation and transfer of disulphide bonds in living cells". Nature Reviews Molecular Cell Biology . 3 (11): 836–847. doi:10.1038/nrm954. PMID 12415301.
- Witt, D. (2008). "Recent developments in disulfide bond formation". Synthesis. 2008 (16): 2491–2509. doi:10.1055/s-2008-1067188.
- M. E. Alonso and H. Aragona (1978). "Sulfide Synthesis in Preparation of Unsymmetrical Dialkyl Disulfides: Sec-butyl Isopropyl Disulfide". Org. Synth. 58: 147. doi:10.15227/orgsyn.058.0147.CS1 maint: uses authors parameter (link)
- TCEP technical information, from Interchim
- Gilbert, H. F. (1990). "Molecular and Cellular Aspects of Thiol–Disulfide Exchange". Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology and Related Areas of Molecular Biology. 63. pp. 69–172. doi:10.1002/9780470123096.ch2. ISBN 9780470123096. PMID 2407068.
- Gilbert, H. F. (1995). "Thiol/disulfide exchange equilibria and disulfide bond stability". Biothiols, Part A: Monothiols and Dithiols, Protein Thiols, and Thiyl Radicals. Methods in Enzymology. 251. pp. 8–28. doi:10.1016/0076-6879(95)51107-5. ISBN 9780121821524. PMID 7651233.
- For example the conversion of di-o-nitrophenyl disulfide to o-nitrophenylsulfur chloride Hubacher, Max H. (1935). "o-Nitrophenylsulfur Chloride". Organic Syntheses. 15: 45. doi:10.15227/orgsyn.015.00452.
- Methyl sulfenyl chloride: Douglass, Irwin B.; Norton, Richard V. (1960). "Methanesulfinyl Chloride". Organic Syntheses. 40: 62. doi:10.15227/orgsyn.040.0062.
- Hatahet, F.; Nguyen, V. D.; Salo, K. E.; Ruddock, L. W. (2010). "Disruption of reducing pathways is not essential for efficient disulfide bond formation in the cytoplasm of E. coli". Microbial Cell Factories. 9 (67): 67. doi:10.1186/1475-2859-9-67. PMC 2946281. PMID 20836848.
- Ruoppolo, M.; Vinci, F.; Klink, T. A.; Raines, R. T.; Marino, G. (2000). "Contribution of individual disulfide bonds to the oxidative folding of ribonuclease A". Biochemistry. 39 (39): 12033–12042. doi:10.1021/bi001044n. PMID 11009618.
- Ladenstein, R.; Ren, B. (2008). "Reconsideration of an early dogma, saying "there is no evidence for disulfide bonds in proteins from archaea"". Extremophiles. 12 (1): 29–38. doi:10.1007/s00792-007-0076-z. PMID 17508126.
- Samson, Andre L.; Knaupp, Anja S.; Sashindranath, Maithili; Borg, Rachael J.; Au, Amanda E.-L.; Cops, Elisa J.; Saunders, Helen M.; Cody, Stephen H.; McLean, Catriona A. (2012-10-25). "Nucleocytoplasmic coagulation: an injury-induced aggregation event that disulfide crosslinks proteins and facilitates their removal by plasmin". Cell Reports. 2 (4): 889–901. doi:10.1016/j.celrep.2012.08.026. ISSN 2211-1247. PMID 23041318.
Further reading
- Sela, M.; Lifson, S. (1959). "The reformation of disulfide bridges in proteins". Biochimica et Biophysica Acta. 36 (2): 471–478. doi:10.1016/0006-3002(59)90188-X. PMID 14444674.
- Stark, G. R.; Stern, K.; Atala, A.; Yoo, J. (1977). "Cleavage at cysteine after cyanylation". Methods in Enzymology. 47 (2): 129–132. doi:10.1016/j.ymeth.2008.09.005. PMID 927170.
- Thornton, J. M. (1981). "Disulphide bridges in globular proteins". Journal of Molecular Biology. 151 (2): 261–287. doi:10.1016/0022-2836(81)90515-5. PMID 7338898.
- Thannhauser, T. W.; Konishi, Y.; Scheraga, H. A. (1984). "Sensitive quantitative analysis of disulfide bonds in polypeptides and proteins". Analytical Biochemistry. 138 (1): 181–188. doi:10.1016/0003-2697(84)90786-3. PMID 6547275.
- Wu, J.; Watson, J. T. (1998). "Optimization of the cleavage reaction for cyanylated cysteinyl proteins for efficient and simplified mass mapping". Analytical Biochemistry. 258 (2): 268–276. doi:10.1006/abio.1998.2596. PMID 9570840.
- Futami, J.; Tada, H.; Seno, M.; Ishikami, S.; Yamada, H. (2000). "Stabilization of human RNAse 1 by introduction of a disulfide bond between residues 4 and 118". Journal of Biochemistry. 128 (2): 245–250. doi:10.1093/oxfordjournals.jbchem.a022747. PMID 10920260.
- Wittenberg, G.; Danon, A. (2008). "Disulfide bond formation in chloroplasts: Formation of disulfide bonds in signaling chloroplast proteins". Plant Science. 175 (4): 459–466. doi:10.1016/j.plantsci.2008.05.011.
- Kadokura, Hiroshi; Katzen, Federico; Beckwith, Jon (2003). "Protein Disulfide Bond Formation in Prokaryotes". Annual Review of Biochemistry. 72 (1): 111–135. doi:10.1146/annurev.biochem.72.121801.161459. PMID 12524212.
- Tu, B. P.; Weissman, J. S. (2004). "Oxidative protein folding in eukaryotes: mechanisms and consequences". The Journal of Cell Biology. 164 (3): 341–346. doi:10.1083/jcb.200311055. PMC 2172237. PMID 14757749.
- Ellgaard, Lars; Ruddock, Lloyd W. (2005). "The human protein disulphide isomerase family: substrate interactions and functional properties". EMBO Reports. 6 (1): 28–32. doi:10.1038/sj.embor.7400311. PMC 1299221. PMID 15643448.
External links