Lysine
Lysine (symbol Lys or K)[1] is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain lysyl ((CH2)4NH2), classifying it as a basic, charged (at physiological pH), aliphatic amino acid. It is encoded by the codons AAA and AAG. Like almost all other amino acids, the α-carbon is chiral and lysine may refer to either enantiomer or a racemic mixture of both. For the purpose of this article, lysine will refer to the biologically active enantiomer L-lysine, where the α-carbon is in the S configuration.
 L-lysine | |
| Names | |
|---|---|
| IUPAC name
(2S)-2,6-Diaminohexanoic acid (L-lysine)
(2R)-2,6-Diaminohexanoic acid (D-lysine) | |
| Other names
Lysine, D-lysine, L-lysine, LYS, h-Lys-OH | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.000.673 |
| KEGG | |
PubChem CID |
|
| UNII | |
| |
| |
| Properties | |
| C6H14N2O2 | |
| Molar mass | 146.190 g·mol−1 |
| 1.5 kg/L | |
| Pharmacology | |
| B05XB03 (WHO) | |
| Supplementary data page | |
| Refractive index (n), Dielectric constant (εr), etc. | |
Thermodynamic data |
Phase behaviour solid–liquid–gas |
| UV, IR, NMR, MS | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
The human body cannot synthesize lysine. It is essential in humans and must be obtained from the diet. In organisms that synthesise lysine, it has two main biosynthetic pathways, the diaminopimelate and α-aminoadipate pathways, which employ different enzymes and substrates and are found in different organisms. Lysine catabolism occurs through one of several pathways, the most common of which is the saccharopine pathway.
Lysine plays several roles in humans, most importantly proteinogenesis, but also in the crosslinking of collagen polypeptides, uptake of essential mineral nutrients, and in the production of carnitine, which is key in fatty acid metabolism. Lysine is also often involved in histone modifications, and thus, impacts the epigenome. The ε-amino group often participates in hydrogen bonding and as a general base in catalysis. The ε-ammonium group (NH3+) is attached to the fourth carbon from the α-carbon, which is attached to the carboxyl (C=OOH) group.[2]
Due to its importance in several biological processes, a lack of lysine can lead to several disease states including defective connective tissues, impaired fatty acid metabolism, anaemia, and systemic protein-energy deficiency. In contrast, an overabundance of lysine, caused by ineffective catabolism, can cause severe neurological disorders.
Lysine was first isolated by the German biological chemist Ferdinand Heinrich Edmund Drechsel in 1889 from the protein casein in milk.[3] He named it "lysin".[4] In 1902, the German chemists Emil Fischer and Fritz Weigert determined lysine's chemical structure by synthesizing it.[5]
Biosynthesis
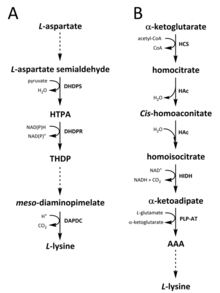
Two different pathways have been identified in nature for the synthesis of lysine. The diaminopimelate (DAP) pathway belongs to the aspartate derived biosynthetic family, which is also involved in the synthesis of threonine, methionine and isoleucine.[6][7] Whereas the α-aminoadipate (AAA) pathway is part of the glutamate biosynthetic family.[8][9]
The DAP pathway is found in both prokaryotes and plants and begins with the dihydrodipicolinate synthase (DHDPS) (E.C 4.3.3.7) catalysed condensation reaction between the aspartate derived, L-aspartate semialdehyde, and pyruvate to form (4S)-4-hydroxy-2,3,4,5-tetrahydro-(2S)-dipicolinic acid (HTPA).[10][11][12][13][14] The product is then reduced by dihydrodipicolinate reductase (DHDPR) (E.C 1.3.1.26), with NAD(P)H as a proton donor, to yield 2,3,4,5-tetrahydrodipicolinate (THDP).[15] From this point on, there are four pathway variations found in different species, namely the acetylase, aminotransferase, dehydrogenase, and succinylase pathways.[6][16] Both the acetylase and succinylase variant pathways use four enzyme catalysed steps, the aminotransferase pathway uses two enzymes, and the dehydrogenase pathway uses a single enzyme.[17] These four variant pathways converge at the formation of the penultimate product, meso‑diaminopimelate, which is subsequently enzymatically decarboxylated in an irreversible reaction catalysed by diaminopimelate decarboxylase (DAPDC) (E.C 4.1.1.20) to produce L-lysine.[18][19] The DAP pathway is regulated at multiple levels, including upstream at the enzymes involved in aspartate processing as well as at the initial DHDPS catalysed condensation step.[19][20] Lysine imparts a strong negative feedback loop on these enzymes and, subsequently, regulates the entire pathway.[20]
The AAA pathway involves the condensation of α-ketoglutarate and acetyl-CoA via the intermediate AAA for the synthesis of L-lysine. This pathway has been shown to be present in several yeast species, as well as protists and higher fungi.[9][21][22][23][24][25][26] It has also been reported that an alternative variant of the AAA route has been found in Thermus thermophilus and Pyrococcus horikoshii, which could indicate that this pathway is more widely spread in prokaryotes than originally proposed.[27][28][29] The first and rate-limiting step in the AAA pathway is the condensation reaction between acetyl-CoA and α‑ketoglutarate catalysed by homocitrate-synthase (HCS) (E.C 2.3.3.14) to give the intermediate homocitryl‑CoA, which is hydrolysed by the same enzyme to produce homocitrate.[30] Homocitrate is enzymatically dehydrated by homoaconitase (HAc) (E.C 4.2.1.36) to yield cis-homoaconitate.[31] HAc then catalyses a second reaction in which cis-homoaconitate undergoes rehydration to produce homoisocitrate.[9] The resulting product undergoes an oxidative decarboxylation by homoisocitrate dehydrogenase (HIDH) (E.C 1.1.1.87) to yield α‑ketoadipate.[9] AAA is then formed via a pyridoxal 5′-phosphate (PLP)-dependent aminotransferase (PLP-AT) (E.C 2.6.1.39), using glutamate as the amino donor.[30] From this point on, the AAA pathway differs depending on the kingdom. In fungi, AAA is reduced to α‑aminoadipate-semialdehyde via AAA reductase (E.C 1.2.1.95) in a unique process involving both adenylation and reduction that is activated by a phosphopantetheinyl transferase (E.C 2.7.8.7).[9] Once the semialdehyde is formed, saccharopine reductase (E.C 1.5.1.10) catalyses a condensation reaction with glutamate and NAD(P)H, as a proton donor, and the imine is reduced to produce the penultimate product, saccharopine.[29] The final step of the pathway in fungi involves the saccharopine dehydrogenase (SDH) (E.C 1.5.1.8) catalysed oxidative deamination of saccharopine, resulting in L-lysine.[9] In a variant AAA pathway found in some prokaryotes, AAA is first converted to N‑acetyl-α-aminoadipate, which is phosphorylated and then reductively dephosphorylated to the ε-aldehyde.[29][30] The aldehyde is then transaminated to N‑acetyl-lysine, which is deacetylated to give L-lysine.[29][30] However, the enzymes involved in this variant pathway need further validation.
Catabolism
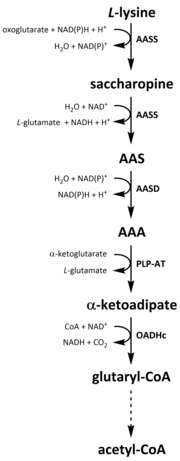
Like all amino acids, catabolism of lysine is initiated from the uptake of dietary lysine or from the breakdown of intracellular protein. Catabolism is also used as a means to control the intracellular concentration of free lysine and maintain a steady-state to prevent the toxic effects of excessive free lysine.[32] There are several pathways involved in lysine catabolism but the most commonly used is the saccharopine pathway, which primarily takes place in the liver (and equivalent organs) in animals, specifically within the mitochondria.[33][32][34][35] This is the reverse of the previously described AAA pathway.[33][36] In animals and plants, the first two steps of the saccharopine pathway are catalysed by the bifunctional enzyme, α-aminoadipic semialdehyde synthase (AASS), which possess both lysine-ketoglutarate reductase (LKR) (E.C 1.5.1.8) and SDH activities, whereas in other organisms, such as bacteria and fungi, both of these enzymes are encoded by separate genes.[37][38] The first step involves the LKR catalysed reduction of L-lysine in the presence of α-ketoglutarate to produce saccharopine, with NAD(P)H acting as a proton donor.[39] Saccharopine then undergoes a dehydration reaction, catalysed by SDH in the presence of NAD+, to produce AAS and glutamate.[40] AAS dehydrogenase (AASD) (E.C 1.2.1.31) then further dehydrates the molecule into AAA.[39] Subsequently, PLP-AT catalyses the reverse reaction to that of the AAA biosynthesis pathway, resulting in AAA being converted to α-ketoadipate. The product, α‑ketoadipate, is decarboxylated in the presence of NAD+ and coenzyme A to yield glutaryl-CoA, however the enzyme involved in this is yet to be fully elucidated.[41][42] Some evidence suggests that the 2-oxoadipate dehydrogenase complex (OADHc), which is structurally homologous to the E1 subunit of the oxoglutarate dehydrogenase complex (OGDHc) (E.C 1.2.4.2), is responsible for the decarboxylation reaction.[41][43] Finally, glutaryl-CoA is oxidatively decarboxylated to crotony-CoA by glutaryl-CoA dehydrogenase (E.C 1.3.8.6), which goes on to be further processed through multiple enzymatic steps to yield acetyl-CoA; an essential carbon metabolite involved in the tricarboxylic acid cycle (TCA).[39][44][45][46]
Nutritional value
Lysine is one of the nine essential amino acids in humans.[47] The human nutritional requirements varies from ~60 mg·kg−1·d−1 in infancy to ~30 mg·kg−1·d−1 in adults.[33] This requirement is commonly met in a western society with the intake of lysine from meat and vegetable sources well in excess of the recommended requirement.[33] In vegetarian diets, the intake of lysine is less due to the limiting quantity of lysine in cereal crops compared to meat sources.[33] Given the limiting concentration of lysine in cereal crops, it has long been speculated that the content of lysine can be increased through genetic modification practices.[48][49] Often these practices have involved the intentional dysregulation of the DAP pathway by means of introducing lysine feedback-insensitive orthologues of the DHDPS enzyme.[48][49] These methods have met limited success likely due to the toxic side effects of increased free lysine and indirect effects on the TCA cycle.[50] Plants accumulate lysine and other amino acids in the form of seed storage proteins, found within the seeds of the plant, and this represents the edible component of cereal crops.[51] This highlights the need to not only increase free lysine, but also direct lysine towards the synthesis of stable seed storage proteins, and subsequently, increase the nutritional value of the consumable component of crops.[52][53] Whilst genetic modification practices have met limited success, more traditional selective breeding techniques have allowed for the isolation of "Quality Protein Maize", which has significantly increased levels of lysine and tryptophan, also an essential amino acid. This increase in lysine content is attributed to an opaque-2 mutation that reduced the transcription of lysine lacking zein related seed storage proteins and, as a result, increased the abundance of other proteins that are rich in lysine.[53][54] Commonly, to overcome the limiting abundance of lysine in livestock feed, industrially produced lysine is added.[55][56] The industrial process includes the fermentative culturing of Corynebacterium glutamicum and the subsequent purification of lysine.[55]
Dietary sources
Good sources of lysine are high-protein foods such as eggs, meat (specifically red meat, lamb, pork, and poultry), soy, beans and peas, cheese (particularly Parmesan), and certain fish (such as cod and sardines).[57] Lysine is the limiting amino acid (the essential amino acid found in the smallest quantity in the particular foodstuff) in most cereal grains, but is plentiful in most pulses (legumes).[58] A vegetarian or low animal protein diet can be adequate for protein, including lysine, if it includes both cereal grains and legumes, but there is no need for the two food groups to be consumed in the same meals.
A food is considered to have sufficient lysine if it has at least 51 mg of lysine per gram of protein (so that the protein is 5.1% lysine).[59] L-lysine HCl is used as a dietary supplement, providing 80.03% L-lysine.[60] As such, 1 g of L-lysine is contained in 1.25 g of L-lysine HCl.
| Food | Lysine (% of protein) |
|---|---|
| Fish | 9.19% |
| Beef, ground, 90% lean/10% fat, cooked | 8.31% |
| Chicken, roasting, meat and skin, cooked, roasted | 8.11% |
| Azuki bean (adzuki beans), mature seeds, raw | 7.53% |
| Milk, non-fat | 7.48% |
| Soybean, mature seeds, raw | 7.42% |
| Egg, whole, raw | 7.27% |
| Pea, split, mature seeds, raw | 7.22% |
| Lentil, pink, raw | 6.97% |
| Kidney bean, mature seeds, raw | 6.87% |
| Chickpea, (garbanzo beans, Bengal gram), mature seeds, raw | 6.69% |
| Navy bean, mature seeds, raw | 5.73% |
Biological roles
The most common role for lysine is proteinogenesis. Lysine frequently plays an important role in protein structure. Since its side chain contains a positively charged group on one end and a long hydrophobic carbon tail close to the backbone, lysine is considered somewhat amphipathic. For this reason, lysine can be found buried as well as more commonly in solvent channels and on the exterior of proteins, where it can interact with the aqueous environment.[61] Lysine can also contribute to protein stability as its ε-amino group often participates in hydrogen bonding, salt bridges and covalent interactions to form a Schiff base.[61][62][63][64]
A second major role of lysine is in epigenetic regulation by means of histone modification.[65][66] There are several types of covalent histone modifications, which commonly involve lysine residues found in the protruding tail of histones. Modifications often include the addition or removal of an acetyl (-CH3CO) forming acetyllysine or reverting to lysine, up to three methyl (‑CH3), ubiquitin or a sumo protein group.[65][67][68][69][70] The various modifications have downstream effects on gene regulation, in which genes can be activated or repressed.
Lysine has also been implicated to play a key role in other biological processes including; structural proteins of connective tissues, calcium homeostasis, and fatty acid metabolism.[71][72][73] Lysine has been shown to be involved in the crosslinking between the three helical polypeptides in collagen, resulting in its stability and tensile strength.[71][74] This mechanism is akin to the role of lysine in bacterial cell walls, in which lysine (and meso-diaminopimelate) are critical to the formation of crosslinks, and therefore, stability of the cell wall.[75] This concept has previously been explored as a means to circumvent the unwanted release of potentially pathogenic genetically modified bacteria. It was proposed that an auxotrophic strain of Escherichia coli (X1776) could be used for all genetic modification practices, as the strain is unable to survive without the supplementation of DAP, and thus, cannot live outside of a laboratory environment.[76] Lysine has also been proposed to be involved in calcium intestinal absorption and renal retention, and thus, may play a role in calcium homeostasis.[72] Finally, lysine has been shown to be a precursor for carnitine, which transports fatty acids to the mitochondria, where they can be oxidised for the release of energy.[73][77] Carnitine is synthesised from trimethyllysine, which is a product of the degradation of certain proteins, as such lysine must first be incorporated into proteins and be methylated prior to being converted to carnitine.[73] However, in mammals the primary source of carnitine is through dietary sources, rather than through lysine conversion.[73]
In opsins like rhodopsin and the visual opsins (encoded by the genes OPN1SW, OPN1MW, and OPN1LW), retinaldehyde forms a Schiff base with a conserved lysine residue, and interaction of light with the retinylidene group causes signal transduction in color vision (See visual cycle for details).
Disputed roles
There has been a long discussion that lysine, when administered intravenously or orally, can significantly increase the release of growth hormones.[78] This has led to athletes using lysine as a means of promoting muscle growth while training, however, no significant evidence to support this application of lysine has been found to date.[78][79]
Because herpes simplex virus (HSV) proteins are richer in arginine and poorer in lysine than the cells they infect, lysine supplements have been tried as a treatment. Since the two amino acids are taken up in the intestine, reclaimed in the kidney, and moved into cells by the same amino acid transporters, an abundance of lysine would, in theory, limit the amount of arginine available for viral replication.[80] Clinical studies do not provide good evidence for effectiveness as a prophylactic or in the treatment for HSV outbreaks.[81][82] In response to product claims that lysine could improve immune responses to HSV, a review by the European Food Safety Authority found no evidence of a cause–effect relationship. The same review, published in 2011, found no evidence to support claims that lysine could lower cholesterol, increase appetite, contribute to protein synthesis in any role other than as an ordinary nutrient, or increase calcium absorption or retention.[83]
Roles in disease
Diseases related to lysine are a result of the downstream processing of lysine, i.e. the incorporation into proteins or modification into alternative biomolecules. The role of lysine in collagen has been outlined above, however, a lack of lysine and hydroxylysine involved in the crosslinking of collagen peptides has been linked to a disease state of the connective tissue.[84] As carnitine is a key lysine-derived metabolite involved in fatty acid metabolism, a substandard diet lacking sufficient carnitine and lysine can lead to decreased carnitine levels, which can have significant cascading effects on an individual's health.[77][85] Lysine has also been shown to play a role in anaemia, as lysine is suspected to have an effect on the uptake of iron and, subsequently, the concentration of ferritin in blood plasma.[86] However, the exact mechanism of action is yet to be elucidated.[86] Most commonly, lysine deficiency is seen in non-western societies and manifests as protein-energy malnutrition, which has profound and systemic effects on the health of the individual.[87][88] There is also a hereditary genetic disease that involves mutations in the enzymes responsible for lysine catabolism, namely the bifunctional AASS enzyme of the saccharopine pathway.[89] Due to a lack of lysine catabolism, the amino acid accumulates in plasma and patients develop hyperlysinaemia, which can present as asymptomatic to severe neurological disabilities, including epilepsy, ataxia, spasticity, and psychomotor impairment.[89][90] The clinical significance of hyperlysinemia is the subject of debate in the field with some studies finding no correlation between physical or mental disabilities and hyperlysinemia.[91] In addition to this, mutations in genes related to lysine metabolism have been implicated in several disease states, including pyridoxine-dependent epilepsia (ALDH7A1 gene), α-ketoadipic and α-aminoadipic aciduria (DHTKD1 gene), and glutaric aciduria type 1 (GCDH gene).[41][92][93][94][95]
Hyperlysinuria is marked by high amounts of lysine in the urine.[96] It is often due to a metabolic disease in which a protein involved in the breakdown of lysine is non functional due to a genetic mutation.[97] It may also occur due to a failure of renal tubular transport.[97]
Use of lysine in animal feed
Lysine production for animal feed is a major global industry, reaching in 2009 almost 700,000 tonnes for a market value of over €1.22 billion.[98] Lysine is an important additive to animal feed because it is a limiting amino acid when optimizing the growth of certain animals such as pigs and chickens for the production of meat. Lysine supplementation allows for the use of lower-cost plant protein (maize, for instance, rather than soy) while maintaining high growth rates, and limiting the pollution from nitrogen excretion.[99] In turn, however, phosphate pollution is a major environmental cost when corn is used as feed for poultry and swine.[100]
Lysine is industrially produced by microbial fermentation, from a base mainly of sugar. Genetic engineering research is actively pursuing bacterial strains to improve the efficiency of production and allow lysine to be made from other substrates.[98]
In popular culture
The 1993 film Jurassic Park (based on the 1990 Michael Crichton novel of the same name) features dinosaurs that were genetically altered so that they could not produce lysine, an example of engineered auxotrophy.[101] This was known as the "lysine contingency" and was supposed to prevent the cloned dinosaurs from surviving outside the park, forcing them to be dependent on lysine supplements provided by the park's veterinary staff. In reality, no animals are capable of producing lysine (it is an essential amino acid).[102]
In 1996, lysine became the focus of a price-fixing case, the largest in United States history. The Archer Daniels Midland Company paid a fine of US$100 million, and three of its executives were convicted and served prison time. Also found guilty in the price-fixing case were two Japanese firms (Ajinomoto, Kyowa Hakko) and a South Korean firm (Sewon).[103] Secret video recordings of the conspirators fixing lysine's price can be found online or by requesting the video from the U.S. Department of Justice, Antitrust Division. This case served as the basis of the movie The Informant!, and a book of the same title.[104]
References
![]()
- "IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature and symbolism for amino acids and peptides. Recommendations 1983". Biochemical Journal. 219 (2): 345–373. 15 April 1984. doi:10.1042/bj2190345. PMC 1153490. PMID 6743224.
- Lysine. The Biology Project, Department of Biochemistry and Molecular Biophysics, University of Arizona.
- Drechsel E (1889). "Zur Kenntniss der Spaltungsprodukte des Caseïns" [[Contribution] to [our] knowledge of the cleavage products of casein]. Journal für Praktische Chemie. 2nd series (in German). 39: 425–429. doi:10.1002/prac.18890390135. On p. 428, Drechsel presented an empirical formula for the chloroplatinate salt of lysine – C8H16N2O2Cl2•PtCl4 + H2O – but he later admitted that this formula was wrong because the salt's crystals contained ethanol instead of water. See: Drechsel E (1891). "Der Abbau der Eiweissstoffe" [The disassembly of proteins]. Archiv für Anatomie und Physiologie (in German): 248–278.; Drechsel E. "Zur Kenntniss der Spaltungsproducte des Caseïns" [Contribution] to [our] knowledge of the cleavage products of casein] (in German): 254–260.
From p. 256:] " … die darin enthaltene Base hat die Formel C6H14N2O2. Der anfängliche Irrthum ist dadurch veranlasst worden, dass das Chloroplatinat nicht, wie angenommen ward, Krystallwasser, sondern Krystallalkohol enthält, … " ( … the base [that's] contained therein has the [empirical] formula C6H14N2O2. The initial error was caused by the chloroplatinate containing not water in the crystal (as was assumed), but ethanol … )
Cite journal requires|journal=(help) - Drechsel E (1891). "Der Abbau der Eiweissstoffe" [The disassembly of proteins]. Archiv für Anatomie und Physiologie (in German): 248–278.; Fischer E (1891). "Ueber neue Spaltungsproducte des Leimes" [On new cleavage products of gelatin] (in German): 465–469.
From p. 469:] " … die Base C6H14N2O2, welche mit dem Namen Lysin bezeichnet werden mag, … " ( … the base C6H14N2O2, which may be designated with the name "lysine", … ) [Note: Ernst Fischer was a graduate student of Drechsel.]
Cite journal requires|journal=(help) - Fischer E, Weigert F (1902). "Synthese der α,ε – Diaminocapronsäure (Inactives Lysin)" [Synthesis of α,ε-diaminohexanoic acid ([optically] inactive lysine)]. Berichte der Deutschen Chemischen Gesellschaft (in German). 35 (3): 3772–3778. doi:10.1002/cber.190203503211.
- Hudson AO, Bless C, Macedo P, Chatterjee SP, Singh BK, Gilvarg C, Leustek T (January 2005). "Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways". Biochimica et Biophysica Acta (BBA) - General Subjects. 1721 (1–3): 27–36. doi:10.1016/j.bbagen.2004.09.008. PMID 15652176.
- Velasco AM, Leguina JI, Lazcano A (October 2002). "Molecular evolution of the lysine biosynthetic pathways". Journal of Molecular Evolution. 55 (4): 445–59. doi:10.1007/s00239-002-2340-2. PMID 12355264.
- Miyazaki T, Miyazaki J, Yamane H, Nishiyama M (July 2004). "alpha-Aminoadipate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus" (PDF). Microbiology. 150 (Pt 7): 2327–34. doi:10.1099/mic.0.27037-0. PMID 15256574.
- Xu H, Andi B, Qian J, West AH, Cook PF (2006). "The alpha-aminoadipate pathway for lysine biosynthesis in fungi". Cell Biochemistry and Biophysics. 46 (1): 43–64. doi:10.1385/CBB:46:1:43. PMID 16943623.
- Atkinson SC, Dogovski C, Downton MT, Czabotar PE, Dobson RC, Gerrard JA, Wagner J, Perugini MA (March 2013). "Structural, kinetic and computational investigation of Vitis vinifera DHDPS reveals new insight into the mechanism of lysine-mediated allosteric inhibition". Plant Molecular Biology. 81 (4–5): 431–46. doi:10.1007/s11103-013-0014-7. PMID 23354837.
- Griffin MD, Billakanti JM, Wason A, Keller S, Mertens HD, Atkinson SC, Dobson RC, Perugini MA, Gerrard JA, Pearce FG (2012). "Characterisation of the first enzymes committed to lysine biosynthesis in Arabidopsis thaliana". PLOS ONE. 7 (7): e40318. doi:10.1371/journal.pone.0040318. PMC 3390394. PMID 22792278.
- Soares da Costa TP, Muscroft-Taylor AC, Dobson RC, Devenish SR, Jameson GB, Gerrard JA (July 2010). "How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?". Biochimie. 92 (7): 837–45. doi:10.1016/j.biochi.2010.03.004. PMID 20353808.
- Soares da Costa TP, Christensen JB, Desbois S, Gordon SE, Gupta R, Hogan CJ, Nelson TG, Downton MT, Gardhi CK, Abbott BM, Wagner J, Panjikar S, Perugini MA (2015). "Quaternary Structure Analyses of an Essential Oligomeric Enzyme". Analytical Ultracentrifugation. Methods in Enzymology. 562. pp. 205–23. doi:10.1016/bs.mie.2015.06.020. ISBN 9780128029084. PMID 26412653.
- Muscroft-Taylor AC, Soares da Costa TP, Gerrard JA (March 2010). "New insights into the mechanism of dihydrodipicolinate synthase using isothermal titration calorimetry". Biochimie. 92 (3): 254–62. doi:10.1016/j.biochi.2009.12.004. PMID 20025926.
- Christensen JB, Soares da Costa TP, Faou P, Pearce FG, Panjikar S, Perugini MA (November 2016). "Structure and Function of Cyanobacterial DHDPS and DHDPR". Scientific Reports. 6 (1): 37111. doi:10.1038/srep37111. PMC 5109050. PMID 27845445.
- McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, Maurelli AT (November 2006). "L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine". Proceedings of the National Academy of Sciences of the United States of America. 103 (47): 17909–14. doi:10.1073/pnas.0608643103. PMC 1693846. PMID 17093042.
- Hudson AO, Gilvarg C, Leustek T (May 2008). "Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of LL-diaminopimelate aminotransferase". Journal of Bacteriology. 190 (9): 3256–63. doi:10.1128/jb.01381-07. PMC 2347407. PMID 18310350.
- Peverelli MG, Perugini MA (August 2015). "An optimized coupled assay for quantifying diaminopimelate decarboxylase activity". Biochimie. 115: 78–85. doi:10.1016/j.biochi.2015.05.004. PMID 25986217.
- Soares da Costa TP, Desbois S, Dogovski C, Gorman MA, Ketaren NE, Paxman JJ, Siddiqui T, Zammit LM, Abbott BM, Robins-Browne RM, Parker MW, Jameson GB, Hall NE, Panjikar S, Perugini MA (August 2016). "Structural Determinants Defining the Allosteric Inhibition of an Essential Antibiotic Target". Structure. 24 (8): 1282–1291. doi:10.1016/j.str.2016.05.019. PMID 27427481.
- Jander G, Joshi V (1 January 2009). "Aspartate-Derived Amino Acid Biosynthesis in Arabidopsis thaliana". The Arabidopsis Book. 7: e0121. doi:10.1199/tab.0121. PMC 3243338. PMID 22303247.
- Andi B, West AH, Cook PF (September 2004). "Kinetic mechanism of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae". Biochemistry. 43 (37): 11790–5. doi:10.1021/bi048766p. PMID 15362863.
- Bhattacharjee JK (1985). "alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes". Critical Reviews in Microbiology. 12 (2): 131–51. doi:10.3109/10408418509104427. PMID 3928261.
- Bhattacharjee JK, Strassman M (May 1967). "Accumulation of tricarboxylic acids related to lysine biosynthesis in a yeast mutant". The Journal of Biological Chemistry. 242 (10): 2542–6. PMID 6026248.
- Gaillardin CM, Ribet AM, Heslot H (November 1982). "Wild-type and mutant forms of homoisocitric dehydrogenase in the yeast Saccharomycopsis lipolytica". European Journal of Biochemistry. 128 (2–3): 489–94. doi:10.1111/j.1432-1033.1982.tb06991.x. PMID 6759120.
- Jaklitsch WM, Kubicek CP (July 1990). "Homocitrate synthase from Penicillium chrysogenum. Localization, purification of the cytosolic isoenzyme, and sensitivity to lysine". The Biochemical Journal. 269 (1): 247–53. doi:10.1042/bj2690247. PMC 1131560. PMID 2115771.
- Ye ZH, Bhattacharjee JK (December 1988). "Lysine biosynthesis pathway and biochemical blocks of lysine auxotrophs of Schizosaccharomyces pombe". Journal of Bacteriology. 170 (12): 5968–70. doi:10.1128/jb.170.12.5968-5970.1988. PMC 211717. PMID 3142867.
- Kobashi N, Nishiyama M, Tanokura M (March 1999). "Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via alpha-aminoadipic acid not via diaminopimelic acid". Journal of Bacteriology. 181 (6): 1713–8. doi:10.1128/JB.181.6.1713-1718.1999. PMC 93567. PMID 10074061.
- Kosuge T, Hoshino T (1999). "The alpha-aminoadipate pathway for lysine biosynthesis is widely distributed among Thermus strains". Journal of Bioscience and Bioengineering. 88 (6): 672–5. doi:10.1016/S1389-1723(00)87099-1. PMID 16232683.
- Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H (December 1999). "A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis". Genome Research. 9 (12): 1175–83. doi:10.1101/gr.9.12.1175. PMID 10613839.
- Nishida H, Nishiyama M (September 2000). "What is characteristic of fungal lysine synthesis through the alpha-aminoadipate pathway?". Journal of Molecular Evolution. 51 (3): 299–302. doi:10.1007/s002390010091. PMID 11029074.
- Zabriskie TM, Jackson MD (February 2000). "Lysine biosynthesis and metabolism in fungi". Natural Product Reports. 17 (1): 85–97. doi:10.1039/a801345d. PMID 10714900.
- Zhu X, Galili G (May 2004). "Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues". Plant Physiology. 135 (1): 129–36. doi:10.1104/pp.103.037168. PMC 429340. PMID 15122025.
- Tomé D, Bos C (June 2007). "Lysine requirement through the human life cycle". The Journal of Nutrition. 137 (6 Suppl 2): 1642S–1645S. doi:10.1093/jn/137.6.1642S. PMID 17513440.
- Blemings KP, Crenshaw TD, Swick RW, Benevenga NJ (August 1994). "Lysine-alpha-ketoglutarate reductase and saccharopine dehydrogenase are located only in the mitochondrial matrix in rat liver". The Journal of Nutrition. 124 (8): 1215–21. doi:10.1093/jn/124.8.1215. PMID 8064371.
- Galili G, Tang G, Zhu X, Gakiere B (June 2001). "Lysine catabolism: a stress and development super-regulated metabolic pathway". Current Opinion in Plant Biology. 4 (3): 261–6. doi:10.1016/s1369-5266(00)00170-9. PMID 11312138.
- Arruda P, Kemper EL, Papes F, Leite A (August 2000). "Regulation of lysine catabolism in higher plants". Trends in Plant Science. 5 (8): 324–30. doi:10.1016/s1360-1385(00)01688-5. PMID 10908876.
- Sacksteder KA, Biery BJ, Morrell JC, Goodman BK, Geisbrecht BV, Cox RP, Gould SJ, Geraghty MT (June 2000). "Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia". American Journal of Human Genetics. 66 (6): 1736–43. doi:10.1086/302919. PMC 1378037. PMID 10775527.
- Zhu X, Tang G, Galili G (December 2002). "The activity of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is regulated by functional interaction between its two enzyme domains". The Journal of Biological Chemistry. 277 (51): 49655–61. doi:10.1074/jbc.m205466200. PMID 12393892.
- Kiyota E, Pena IA, Arruda P (November 2015). "The saccharopine pathway in seed development and stress response of maize". Plant, Cell & Environment. 38 (11): 2450–61. doi:10.1111/pce.12563. PMID 25929294.
- Serrano GC, Rezende e Silva Figueira T, Kiyota E, Zanata N, Arruda P (March 2012). "Lysine degradation through the saccharopine pathway in bacteria: LKR and SDH in bacteria and its relationship to the plant and animal enzymes". FEBS Letters. 586 (6): 905–11. doi:10.1016/j.febslet.2012.02.023. PMID 22449979.
- Danhauser K, Sauer SW, Haack TB, Wieland T, Staufner C, Graf E, Zschocke J, Strom TM, Traub T, Okun JG, Meitinger T, Hoffmann GF, Prokisch H, Kölker S (December 2012). "DHTKD1 mutations cause 2-aminoadipic and 2-oxoadipic aciduria". American Journal of Human Genetics. 91 (6): 1082–7. doi:10.1016/j.ajhg.2012.10.006. PMC 3516599. PMID 23141293.
- Sauer SW, Opp S, Hoffmann GF, Koeller DM, Okun JG, Kölker S (January 2011). "Therapeutic modulation of cerebral L-lysine metabolism in a mouse model for glutaric aciduria type I". Brain. 134 (Pt 1): 157–70. doi:10.1093/brain/awq269. PMID 20923787.
- Goncalves RL, Bunik VI, Brand MD (February 2016). "Production of superoxide/hydrogen peroxide by the mitochondrial 2-oxoadipate dehydrogenase complex". Free Radical Biology & Medicine. 91: 247–55. doi:10.1016/j.freeradbiomed.2015.12.020. PMID 26708453.
- Goh DL, Patel A, Thomas GH, Salomons GS, Schor DS, Jakobs C, Geraghty MT (July 2002). "Characterization of the human gene encoding alpha-aminoadipate aminotransferase (AADAT)". Molecular Genetics and Metabolism. 76 (3): 172–80. doi:10.1016/s1096-7192(02)00037-9. PMID 12126930.
- Härtel U, Eckel E, Koch J, Fuchs G, Linder D, Buckel W (1 February 1993). "Purification of glutaryl-CoA dehydrogenase from Pseudomonas sp., an enzyme involved in the anaerobic degradation of benzoate". Archives of Microbiology. 159 (2): 174–81. doi:10.1007/bf00250279. PMID 8439237.
- Sauer SW (October 2007). "Biochemistry and bioenergetics of glutaryl-CoA dehydrogenase deficiency". Journal of Inherited Metabolic Disease. 30 (5): 673–80. doi:10.1007/s10545-007-0678-8. PMID 17879145.
- Nelson DL, Cox MM, Lehninger AL (2013). Lehninger principles of biochemistry (6th ed.). New York: W.H. Freeman and Company. ISBN 978-1-4641-0962-1. OCLC 824794893.
- Galili G, Amir R (February 2013). "Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality". Plant Biotechnology Journal. 11 (2): 211–22. doi:10.1111/pbi.12025. PMID 23279001.
- Wang G, Xu M, Wang W, Galili G (June 2017). "Fortifying Horticultural Crops with Essential Amino Acids: A Review". International Journal of Molecular Sciences. 18 (6): 1306. doi:10.3390/ijms18061306. PMC 5486127. PMID 28629176.
- Angelovici R, Fait A, Fernie AR, Galili G (January 2011). "A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination". The New Phytologist. 189 (1): 148–59. doi:10.1111/j.1469-8137.2010.03478.x. PMID 20946418.
- Edelman M, Colt M (2016). "Nutrient Value of Leaf vs. Seed". Frontiers in Chemistry. 4: 32. doi:10.3389/fchem.2016.00032. PMC 4954856. PMID 27493937.
- Jiang SY, Ma A, Xie L, Ramachandran S (September 2016). "Improving protein content and quality by over-expressing artificially synthetic fusion proteins with high lysine and threonine constituent in rice plants". Scientific Reports. 6 (1): 34427. doi:10.1038/srep34427. PMC 5039639. PMID 27677708.
- Shewry PR (November 2007). "Improving the protein content and composition of cereal grain". Journal of Cereal Science. 46 (3): 239–250. doi:10.1016/j.jcs.2007.06.006.
- Prasanna B, Vasal SK, Kassahun B, Singh NN (2001). "Quality protein maize". Current Science. 81 (10): 1308–1319. JSTOR 24105845.
- Kircher M, Pfefferle W (April 2001). "The fermentative production of L-lysine as an animal feed additive". Chemosphere. 43 (1): 27–31. doi:10.1016/s0045-6535(00)00320-9. PMID 11233822.
- Junior L, Alberto L, Letti GV, Soccol CR, Junior L, Alberto L, Letti GV, Soccol CR (2016). "Development of an L-Lysine Enriched Bran for Animal Nutrition via Submerged Fermentation by Corynebacterium glutamicum using Agroindustrial Substrates". Brazilian Archives of Biology and Technology. 59. doi:10.1590/1678-4324-2016150519. ISSN 1516-8913.
- University of Maryland Medical Center. "Lysine". Retrieved 30 December 2009.
- Young VR, Pellett PL (1994). "Plant proteins in relation to human protein and amino acid nutrition". American Journal of Clinical Nutrition. 59 (5 Suppl): 1203S–1212S. doi:10.1093/ajcn/59.5.1203s. PMID 8172124.
- Institute of Medicine of the National Academies. "Dietary Reference Intakes for Macronutrients". p. 589. Retrieved 29 October 2017.
- "Dietary Supplement Database: Blend Information (DSBI)".
L-LYSINE HCL 10000820 80.03% lysine
- Betts MJ, Russell RB (2003). Barnes MR, Gray IC (eds.). Bioinformatics for Geneticists. John Wiley & Sons, Ltd. pp. 289–316. doi:10.1002/0470867302.ch14. ISBN 978-0-470-86730-3.
- Blickling S, Renner C, Laber B, Pohlenz HD, Holak TA, Huber R (January 1997). "Reaction mechanism of Escherichia coli dihydrodipicolinate synthase investigated by X-ray crystallography and NMR spectroscopy". Biochemistry. 36 (1): 24–33. doi:10.1021/bi962272d. PMID 8993314.
- Kumar S, Tsai CJ, Nussinov R (March 2000). "Factors enhancing protein thermostability". Protein Engineering. 13 (3): 179–91. doi:10.1093/protein/13.3.179. PMID 10775659.
- Sokalingam S, Raghunathan G, Soundrarajan N, Lee SG (9 July 2012). "A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein". PLOS ONE. 7 (7): e40410. doi:10.1371/journal.pone.0040410. PMC 3392243. PMID 22792305.
- Dambacher S, Hahn M, Schotta G (July 2010). "Epigenetic regulation of development by histone lysine methylation". Heredity. 105 (1): 24–37. doi:10.1038/hdy.2010.49. PMID 20442736.
- Martin C, Zhang Y (November 2005). "The diverse functions of histone lysine methylation". Nature Reviews. Molecular Cell Biology. 6 (11): 838–49. doi:10.1038/nrm1761. PMID 16261189.
- Black JC, Van Rechem C, Whetstine JR (November 2012). "Histone lysine methylation dynamics: establishment, regulation, and biological impact". Molecular Cell. 48 (4): 491–507. doi:10.1016/j.molcel.2012.11.006. PMC 3861058. PMID 23200123.
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (August 2009). "Lysine acetylation targets protein complexes and co-regulates major cellular functions". Science. 325 (5942): 834–40. doi:10.1126/science.1175371. PMID 19608861.
- Shiio Y, Eisenman RN (November 2003). "Histone sumoylation is associated with transcriptional repression". Proceedings of the National Academy of Sciences of the United States of America. 100 (23): 13225–30. doi:10.1073/pnas.1735528100. PMC 263760. PMID 14578449.
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (October 2004). "Role of histone H2A ubiquitination in Polycomb silencing". Nature. 431 (7010): 873–8. doi:10.1038/nature02985. hdl:10261/73732. PMID 15386022.
- Shoulders MD, Raines RT (2009). "Collagen structure and stability". Annual Review of Biochemistry. 78: 929–58. doi:10.1146/annurev.biochem.77.032207.120833. PMC 2846778. PMID 19344236.
- Civitelli R, Villareal DT, Agnusdei D, Nardi P, Avioli LV, Gennari C (1992). "Dietary L-lysine and calcium metabolism in humans". Nutrition. 8 (6): 400–5. PMID 1486246.
- Vaz FM, Wanders RJ (February 2002). "Carnitine biosynthesis in mammals". The Biochemical Journal. 361 (Pt 3): 417–29. doi:10.1042/bj3610417. PMC 1222323. PMID 11802770.
- Yamauchi M, Sricholpech M (25 May 2012). "Lysine post-translational modifications of collagen". Essays in Biochemistry. 52: 113–33. doi:10.1042/bse0520113. PMC 3499978. PMID 22708567.
- Vollmer W, Blanot D, de Pedro MA (March 2008). "Peptidoglycan structure and architecture". FEMS Microbiology Reviews. 32 (2): 149–67. doi:10.1111/j.1574-6976.2007.00094.x. PMID 18194336.
- Curtiss R (May 1978). "Biological containment and cloning vector transmissibility". The Journal of Infectious Diseases. 137 (5): 668–75. doi:10.1093/infdis/137.5.668. PMID 351084.
- Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q (April 2010). "Role of carnitine in disease". Nutrition & Metabolism. 7: 30. doi:10.1186/1743-7075-7-30. PMC 2861661. PMID 20398344.
- Chromiak JA, Antonio J (2002). "Use of amino acids as growth hormone-releasing agents by athletes". Nutrition. 18 (7–8): 657–61. doi:10.1016/s0899-9007(02)00807-9. PMID 12093449.
- Corpas E, Blackman MR, Roberson R, Scholfield D, Harman SM (July 1993). "Oral arginine-lysine does not increase growth hormone or insulin-like growth factor-I in old men". Journal of Gerontology. 48 (4): M128–33. doi:10.1093/geronj/48.4.M128. PMID 8315224.
- Gaby AR (2006). "Natural remedies for Herpes simplex". Altern Med Rev. 11 (2): 93–101. PMID 16813459.
- Tomblin FA, Lucas KH (2001). "Lysine for management of herpes labialis". Am J Health Syst Pharm. 58 (4): 298–300, 304. doi:10.1093/ajhp/58.4.298. PMID 11225166.
- Chi CC, Wang SH, Delamere FM, Wojnarowska F, Peters MC, Kanjirath PP (7 August 2015). "Interventions for prevention of herpes simplex labialis (cold sores on the lips)". The Cochrane Database of Systematic Reviews (8): CD010095. doi:10.1002/14651858.CD010095.pub2. PMC 6461191. PMID 26252373.
- "Scientific Opinion on the substantiation of health claims related to L-lysine and immune defence against herpes virus (ID 453), maintenance of normal blood LDL-cholesterol concentrations (ID 454, 4669), increase in appetite leading to an increase in energ". EFSA Journal. 9 (4): 2063. 2011. doi:10.2903/j.efsa.2011.2063. ISSN 1831-4732.
- Pinnell SR, Krane SM, Kenzora JE, Glimcher MJ (May 1972). "A heritable disorder of connective tissue. Hydroxylysine-deficient collagen disease". The New England Journal of Medicine. 286 (19): 1013–20. doi:10.1056/NEJM197205112861901. PMID 5016372.
- Rudman D, Sewell CW, Ansley JD (September 1977). "Deficiency of carnitine in cachectic cirrhotic patients". The Journal of Clinical Investigation. 60 (3): 716–23. doi:10.1172/jci108824. PMC 372417. PMID 893675.
- Rushton DH (July 2002). "Nutritional factors and hair loss". Clinical and Experimental Dermatology. 27 (5): 396–404. doi:10.1046/j.1365-2230.2002.01076.x. PMID 12190640.
- Emery PW (October 2005). "Metabolic changes in malnutrition". Eye. 19 (10): 1029–34. doi:10.1038/sj.eye.6701959. PMID 16304580.
- Ghosh S, Smriga M, Vuvor F, Suri D, Mohammed H, Armah SM, Scrimshaw NS (October 2010). "Effect of lysine supplementation on health and morbidity in subjects belonging to poor peri-urban households in Accra, Ghana". The American Journal of Clinical Nutrition. 92 (4): 928–39. doi:10.3945/ajcn.2009.28834. PMID 20720257.
- Houten SM, Te Brinke H, Denis S, Ruiter JP, Knegt AC, de Klerk JB, Augoustides-Savvopoulou P, Häberle J, Baumgartner MR, Coşkun T, Zschocke J, Sass JO, Poll-The BT, Wanders RJ, Duran M (April 2013). "Genetic basis of hyperlysinemia". Orphanet Journal of Rare Diseases. 8: 57. doi:10.1186/1750-1172-8-57. PMC 3626681. PMID 23570448.
- Hoffmann GF, Kölker S (2016). Inborn Metabolic Diseases. Springer, Berlin, Heidelberg. pp. 333–348. doi:10.1007/978-3-662-49771-5_22. ISBN 978-3-662-49769-2.
- Dancis J, Hutzler J, Ampola MG, Shih VE, van Gelderen HH, Kirby LT, Woody NC (May 1983). "The prognosis of hyperlysinemia: an interim report". American Journal of Human Genetics. 35 (3): 438–42. PMC 1685659. PMID 6407303.
- Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MA, Omran H, Tacke U, Uhlenberg B, Weschke B, Clayton PT (March 2006). "Mutations in antiquitin in individuals with pyridoxine-dependent seizures". Nature Medicine. 12 (3): 307–9. doi:10.1038/nm1366. PMID 16491085.
- Mills PB, Footitt EJ, Mills KA, Tuschl K, Aylett S, Varadkar S, Hemingway C, Marlow N, Rennie J, Baxter P, Dulac O, Nabbout R, Craigen WJ, Schmitt B, Feillet F, Christensen E, De Lonlay P, Pike MG, Hughes MI, Struys EA, Jakobs C, Zuberi SM, Clayton PT (July 2010). "Genotypic and phenotypic spectrum of pyridoxine-dependent epilepsy (ALDH7A1 deficiency)". Brain. 133 (Pt 7): 2148–59. doi:10.1093/brain/awq143. PMC 2892945. PMID 20554659.
- Hagen J, te Brinke H, Wanders RJ, Knegt AC, Oussoren E, Hoogeboom AJ, Ruijter GJ, Becker D, Schwab KO, Franke I, Duran M, Waterham HR, Sass JO, Houten SM (September 2015). "Genetic basis of alpha-aminoadipic and alpha-ketoadipic aciduria". Journal of Inherited Metabolic Disease. 38 (5): 873–9. doi:10.1007/s10545-015-9841-9. PMID 25860818.
- Hedlund GL, Longo N, Pasquali M (May 2006). "Glutaric acidemia type 1". American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 142C (2): 86–94. doi:10.1002/ajmg.c.30088. PMC 2556991. PMID 16602100.
- "Hyperlysinuria | Define Hyperlysinuria at Dictionary.com".
- Walter, John; John Fernandes; Jean-Marie Saudubray; Georges van den Berghe (2006). Inborn Metabolic Diseases: Diagnosis and Treatment. Berlin: Springer. p. 296. ISBN 978-3-540-28783-4.
- "Norwegian granted for improving lysine production process". All About Feed. 26 January 2010. Archived from the original on 11 March 2012.
- Toride Y (2004). "Lysine and other amino acids for feed: production and contribution to protein utilization in animal feeding". Protein sources for the animal feed industry; FAO Expert Consultation and Workshop on Protein Sources for the Animal Feed Industry; Bangkok, 29 April - 3 May 2002. Rome: Food and Agriculture Organization of the United Nations. ISBN 978-92-5-105012-5.
- Abelson PH (March 1999). "A potential phosphate crisis". Science. 283 (5410): 2015. doi:10.1126/science.283.5410.2015. PMID 10206902.
- Coyne JA (10 October 1999). "The Truth Is Way Out There". The New York Times. Retrieved 6 April 2008.
- Wu G (May 2009). "Amino acids: metabolism, functions, and nutrition". Amino Acids. 37 (1): 1–17. doi:10.1007/s00726-009-0269-0. PMID 19301095.
- Connor JM (2008). Global Price Fixing (2nd ed.). Heidelberg: Springer-Verlag. ISBN 978-3-540-78669-6.
- Eichenwald K (2000). The Informant: a true story. New York: Broadway Books. ISBN 978-0-7679-0326-4.
