Tetratricopeptide repeat
The tetratricopeptide repeat (TPR) is a structural motif. It consists of a degenerate 34 amino acid tandem repeat identified in a wide variety of proteins. It is found in tandem arrays of 3–16 motifs,[1] which form scaffolds to mediate protein–protein interactions and often the assembly of multiprotein complexes. These alpha-helix pair repeats usually fold together to produce a single, linear solenoid domain called a TPR domain. Proteins with such domains include the anaphase-promoting complex (APC) subunits cdc16, cdc23 and cdc27, the NADPH oxidase subunit p67-phox, hsp90-binding immunophilins, transcription factors, the protein kinase R (PKR), the major receptor for peroxisomal matrix protein import PEX5, protein arginine methyltransferase 9 (PRMT9), and mitochondrial import proteins.
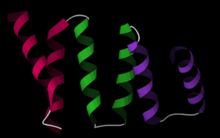
| Tetratricopeptide repeat | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | TPR_1 | ||||||||||
| Pfam | PF00515 | ||||||||||
| Pfam clan | CL0020 | ||||||||||
| InterPro | IPR001440 | ||||||||||
| SCOPe | 1a17 / SUPFAM | ||||||||||
| CDD | cd00189 | ||||||||||
| |||||||||||
Structure
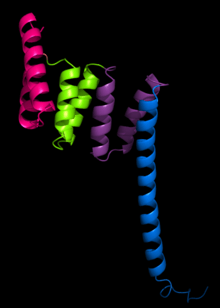
The structure of the PP5 protein was the first structure to be determined. The structure solved by X-ray crystallography by Das and colleagues showed that the TPR sequence motif was composed of a pair of antiparallel alpha helices.[2] The PP5 structure contained 3 tandem TPR repeats which showed the sequential TPR repeats formed an alpha-helical solenoid structure.
A typical TPR structure is characterized by interactions between helices A and B of the first motif and helix A’ of the next TPR. Although the nature of such interactions may vary, the first two helices of the TPR motif typically have a packing angle of ~24 degrees within a single motif. Repeats of more than three TPR motifs generate a right handed superhelix characterized by both a concave and a convex face, of which the concave face is usually involved in ligand binding.[1] [3]
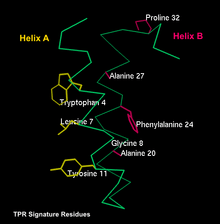
In terms of sequence, a TPR possesses a mixture of small and large hydrophobic residues, nonetheless, no positions are fully invariant. There are however certain residues that are usually conserved including Tryptophan 4, Leucine 7, Glycine 8, Tyrosine 11, Alanine 20, Phenylalanine 24, Alanine 27 and Proline 32. Among those 8, Alanine at positions 8, 20 and 27 tend to be more conserved. The other positions have a stronger preference for either small, large or aromatic amino acids rather than a specific residue. In between helices, residue conservation plays more of a structural role with helix breaking residues present. Between adjacent TPR, residues have roles with both structural and functional implications.[1]
TPR containing peptides
Hop
The Hop adaptor protein mediates the association of the molecular chaperones Hsp70 and Hsp90. It contains three 3-TPR repeats each with its own peptide-binding specificity. Its TPR1 domain is known to recognize the C-terminal of Hsp70 while TPR2 binds to the C-terminal of Hsp90. Both C-terminal sequences end with an EEVD motif and the nature of the interaction is both electrostatic and hydrophobic.[1][4]
PEX5
The PEX5 protein is a receptor for PTS1 (peroxisomal targeting signal tripeptide which directs proteins into peroxisomes). It interacts with the signal via TPR motifs. Most of its contacts with the C-terminal tripeptide PTS1 are in the concave face of TPRs 1, 2 and 3.[5]
Neutrophil cytosolic factor 2
Neutrophil cytosolic factor 2 is an essential to NADPH oxidase complex which in turn produces superoxides in response to microbial infection. The binding of the Rac GTPase is a key step into the assembly of the complex and the TPRs in the phox unit mediate the assembly of the multiprotein complex by acting a binding scaffold.[6]
Examples
Human genes encoding proteins containing this motif include:
- AAG2, ANAPC7
- BBS4
- CABIN1, CDC16, CDC23, CDC27, CNOT10, CTR9
- DNAJC3, DNAJC7, DYX1C1
- FAM10A4, FAM10A5, FKBP4, FKBP5, FKBP8, FKBPL
- GPSM1, GPSM2, GTF3C3
- IFIT1, IFIT1L, IFIT2, IFIT3, IFIT5, IFT140, IFT88
- KLC1, KLC2, KLC3, KLC4, KNS2
- LONRF2
- NARG1, NARG1L, NASP, NCF2, NFKBIL2, NOXA1, NPHP3
- OGT
- PEX5, PEX5L, PPID, PPP5C, PRPF6
- RANBP2, RANBP2L2, RANBP2L6, RAPSN, RGPD5, RGPD7, RPAP3
- SGTA, SGTB, SH3TC1, SH3TC2, SPAG1, SRP72, ST13, STIP1, STUB1, SUGT1
- TMTC1, TMTC2, TMTC3, TMTC4, TOMM34, TOMM70A
- TTC1, TTC3, TTC4, TTC5, TTC6, TTC7A, TTC7B, TTC8, TTC9C, TTC12, TTC13, TTC14, TTC15, TTC16, TTC17, TTC18, TTC21A, TTC21B, TTC22, TTC24, TTC25, TTC27, TTC28, TTC29, TTC30A, TTC30B, TTC31, TTC33, TTC37
- UNC45A, UNC45B, UTX, UTY
- WDTC1
- ZC3H7B
References
- Blatch GL, Lässle M (Nov 1999). "The tetratricopeptide repeat: a structural motif mediating protein-protein interactions". BioEssays. 21 (11): 932–9. doi:10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. PMID 10517866.
- Das AK, Cohen PW, Barford D (Mar 1998). "The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions". The EMBO Journal. 17 (5): 1192–9. doi:10.1093/emboj/17.5.1192. PMC 1170467. PMID 9482716.
- Wilson CG, Kajander T, Regan L (Jan 2005). "The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold". The FEBS Journal. 272 (1): 166–79. doi:10.1111/j.1432-1033.2004.04397.x. PMID 15634341.
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I (Apr 2000). "Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine". Cell. 101 (2): 199–210. doi:10.1016/S0092-8674(00)80830-2. PMID 10786835.
- Gatto GJ, Geisbrecht BV, Gould SJ, Berg JM (Dec 2000). "Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5". Nature Structural Biology. 7 (12): 1091–5. doi:10.1038/81930. PMID 11101887.
- Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K (Oct 2000). "Structure of the TPR domain of p67phox in complex with Rac.GTP". Molecular Cell. 6 (4): 899–907. doi:10.1016/S1097-2765(05)00091-2. PMID 11090627.
Further reading
- Lima Mde F, Eloy NB, Pegoraro C, Sagit R, Rojas C, Bretz T, Vargas L, Elofsson A, de Oliveira AC, Hemerly AS, Ferreira PC (Nov 18, 2010). "Genomic evolution and complexity of the Anaphase-promoting Complex (APC) in land plants". BMC Plant Biology. 10: 254. doi:10.1186/1471-2229-10-254. PMC 3095333. PMID 21087491.
- Das AK, Cohen PW, Barford D (Mar 1998). "The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions". The EMBO Journal. 17 (5): 1192–9. doi:10.1093/emboj/17.5.1192. PMC 1170467. PMID 9482716.
- Whitfield C, Mainprize IL (Feb 2010). "TPR motifs: hallmarks of a new polysaccharide export scaffold". Structure. 18 (2): 151–3. doi:10.1016/j.str.2010.01.006. PMID 20159460.
- Krachler AM, Sharma A, Kleanthous C (Jul 2010). "Self-association of TPR domains: Lessons learned from a designed, consensus-based TPR oligomer". Proteins. 78 (9): 2131–43. doi:10.1002/prot.22726. PMID 20455268.
- Schapire AL, Valpuesta V, Botella MA (Sep 2006). "TPR Proteins in Plant Hormone Signaling". Plant Signaling & Behavior. 1 (5): 229–30. doi:10.4161/psb.1.5.3491. PMC 2634123. PMID 19704665.
- Cortajarena AL, Regan L (May 2006). "Ligand binding by TPR domains". Protein Science. 15 (5): 1193–8. doi:10.1110/ps.062092506. PMC 2242500. PMID 16641492.
- D'Andrea LD, Regan L (Dec 2003). "TPR proteins: the versatile helix". Trends in Biochemical Sciences. 28 (12): 655–62. CiteSeerX 10.1.1.313.8712. doi:10.1016/j.tibs.2003.10.007. PMID 14659697.
- Goebl M, Yanagida M (May 1991). "The TPR snap helix: a novel protein repeat motif from mitosis to transcription". Trends in Biochemical Sciences. 16 (5): 173–7. doi:10.1016/0968-0004(91)90070-C. PMID 1882418.
External links
- Eukaryotic Linear Motif resource motif class LIG_TPR
- Eukaryotic Linear Motif resource motif class TRG_PTS1