Pickover stalk
Pickover stalks are certain kinds of details to be found empirically in the Mandelbrot set, in the study of fractal geometry.[1] They are so named after the researcher Clifford Pickover, whose "epsilon cross" method was instrumental in their discovery. An "epsilon cross" is a cross-shaped orbit trap.

According to Vepstas (1997) "Pickover hit on the novel concept of looking to see how closely the orbits of interior points come to the x and y axes. In these pictures, the closer that the point approaches, the higher up the color scale, with red denoting the closest approach. The logarithm of the distance is taken to accentuate the details".[2]
Biomorphs

Biomorphs are biological-looking Pickover Stalks. [3] At the end of the 1980s, Pickover developed biological feedback organisms similar to Julia sets and the fractal Mandelbrot set.[4] According to Pickover (1999) in summary, he "described an algorithm which could be used for the creation of diverse and complicated forms resembling invertebrate organisms. The shapes are complicated and difficult to predict before actually experimenting with the mappings. He hoped these techniques would encourage others to explore further and discover new forms, by accident, that are on the edge of science and art".[5]
Pickover developed an algorithm (which uses neither random perturbations nor natural laws) to create very complicated forms resembling invertebrate organisms. The iteration, or recursion, of mathematical transformations is used to generate biological morphologies. He called them "biomorphs." At the same time he coined "biomorph" for these patterns, the famous evolutionary biologist Richard Dawkins used the word to refer to his own set of biological shapes that were arrived at by a very different procedure. More rigorously, Pickover's "biomorphs" encompass the class of organismic morphologies created by small changes to traditional convergence tests in the field of "Julia set" theory.[5]
Pickover's biomorphs show a self-similarity at different scales, a common feature of dynamical systems with feedback. Real systems, such as shorelines and mountain ranges, also show self-similarity over some scales. A 2-dimensional parametric 0L system can “look” like Pickover's biomorphs.[6]
Implementation
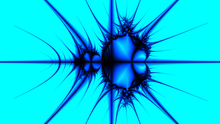
The below example, written in pseudocode, renders a Mandelbrot set colored using a Pickover Stalk with a transformation vector and a color dividend.
The transformation vector is used to offset the (x, y) position when sampling the point's distance to the horizontal and vertical axis.
The color dividend is a float used to determine how thick the stalk is when it is rendered.
For each pixel (x, y) on the target, do:
{
zx = scaled x coordinate of pixel (scaled to lie in the Mandelbrot X scale (-2.5, 1))
zy = scaled y coordinate of pixel (scaled to lie in the Mandelbrot Y scale (-1, 1))
float2 c = (zx, zy) //Offset in the Mandelbrot formulae
float x = zx; //Coordinates to be iterated
float y = zy;
float trapDistance = 1000000; //Keeps track of distance, set to a high value at first.
int iteration = 0;
while (x*x + y*y < 4 && iteration < maxIterations)
{
float2 z = float2(x, y);
z = cmul(z, z); // z^2, cmul is a multiplication function for complex numbers
z += c;
x = z.x;
y = z.y;
float distanceToX = abs(z.x + transformationVector.x); //Checks the distance to the vertical axis
float distanceToY = abs(z.y + transformationVector.y); //Checks the distance to the horizontal axis
smallestDistance = min(distanceToX, distanceToY); // Use only smaller axis distance
trapDistance = min(trapDistance, smallestDistance);
iteration++;
}
return trapDistance * color / dividend;
//Dividend is an external float, the higher it is the thicker the stalk is
}
References
- Peter J. Bentley and David W. Corne (2001). Creative Evolutionary Systems. Morgan Kaufmann. p. 354.
- Linas Vepstas (1997). "Interior Sketchbook Diary". Retrieved 8 July 2008.
- Paul Nylander. Mandelbrot Set Biomorph. feb 2005. Retrieved 8 July 2008.
- Edward Rietman (1994). Genesis Redux: Experiments Creating Artificial Life. Windcrest/McGraw-Hill. p. 154.
- Clifford A. Pickover (1991) "Accident, Evolution, and Art". YLEN NEWSLETTER number. 12 volume 19 Nov/Dec. 1999.
- Alfonso Ortega, Marina de la Cruz, and Manuel Alfonseca (2002). "Parametric 2-dimensional L systems and recursive fractal images: Mandelbrot set, Julia sets and biomorphs". In: Computers & Graphics Volume 26, Issue 1, February 2002, Pages 143-149.
Further reading
- Pickover, Clifford (1987). "Biomorphs: Computer Displays of Biological Forms Generated from Mathematical Feedback Loops". Computer Graphics Forum. 5 (4): 313–316. doi:10.1111/j.1467-8659.1986.tb00317.x.
External links
- Apeirographic Explorations: Biomorphs A random assortment of biomorphs.
- Mad Teddy's Biomorphs, detailed write-up on Pickover's algorithm, including examples and source code.





.jpg)
.jpg)