Phytanoyl-CoA dioxygenase
In enzymology, a phytanoyl-CoA dioxygenase (EC 1.14.11.18) is an enzyme that catalyzes the chemical reaction
- phytanoyl-CoA + 2-oxoglutarate + O2 2-hydroxyphytanoyl-CoA + succinate + CO2
| phytanoyl-CoA dioxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.14.11.18 | ||||||||
| CAS number | 185402-46-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| phytanoyl-CoA 2-hydroxylase | |
|---|---|
| Identifiers | |
| Symbol | PHYH |
| Alt. symbols | PAHX |
| NCBI gene | 5264 |
| HGNC | 8940 |
| OMIM | 602026 |
| RefSeq | NM_001037537 |
| UniProt | O14832 |
| Other data | |
| Locus | Chr. 10 p15.3-10p12.2 |
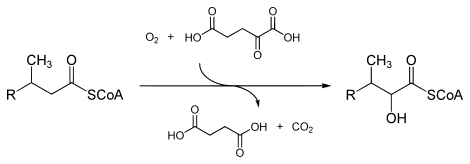
The three substrates of this enzyme are phytanoyl-CoA, 2-oxoglutarate (2OG), and O2, whereas its three products are 2-hydroxyphytanoyl-CoA, succinate, and CO2.
This enzyme belongs to the family of iron(II)-dependent oxygenases, which typically incorporate one atom of dioxygen into the substrate and one atom into the succinate carboxylate group. The mechanism is complex, but is believed to involve ordered binding of 2-oxoglutarate to the iron(II) containing enzyme followed by substrate. Binding of substrate causes displacement of a water molecule from the iron(II) cofactor, leaving a vacant coordination position to which dioxygen binds. A rearrangement occurs to form a high energy iron-oxygen species (which is generally thought to be an iron(IV)=O species) that performs the actual oxidation reaction.[2][3]
Nomenclature
The systematic name of this enzyme class is phytanoyl-CoA, 2-oxoglutarate:oxygen oxidoreductase (2-hydroxylating). This enzyme is also called phytanoyl-CoA hydroxylase and phytanoyl-CoA alpha-hydroxylase.[4]
Examples
In humans, phytanoyl-CoA hydroxylase is encoded by the PHYH (aka PAHX) gene and is required for the alpha-oxidation of branched chain fatty acids (e.g. phytanic acid) in peroxisomes. PHYH deficiency results in the accumulation of large tissue stores of phytanic acid and is the major cause of Refsum disease.[5]
Related enzymes
Iron(II) and 2OG-dependent oxygenases are common in microorganisms, plants, and animals; the human genome is predicted to contain about 80 examples, and the model plant Arabidopsis thaliana likely contains more.[2] In plants and microorganisms this enzyme family is associated with a large diversity of oxidative reactions.[6]
References
- McDonough MA, Kavanagh KL, Butler D, Searls T, Oppermann U, Schofield CJ (Dec 2005). "Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease". The Journal of Biological Chemistry. 280 (49): 41101–10. doi:10.1074/jbc.M507528200. PMID 16186124.
- Hausinger, Robert P. (2015). "CHAPTER 1. Biochemical Diversity of 2-Oxoglutarate-Dependent Oxygenases". 2-Oxoglutarate-Dependent Oxygenases. Metallobiology. pp. 1–58. doi:10.1039/9781782621959-00001. ISBN 978-1-84973-950-4.
- Martinez S, Hausinger RP (Aug 2015). "Catalytic Mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases". The Journal of Biological Chemistry. 290 (34): 20702–11. doi:10.1074/jbc.R115.648691. PMC 4543632. PMID 26152721.
- "PHYH phytanoyl-CoA 2-hydroxylase [ Homo sapiens (human) ]". National Center for Biotechnology Information.
- Mihalik SJ, Morrell JC, Kim D, Sacksteder KA, Watkins PA, Gould SJ (Oct 1997). "Identification of PAHX, a Refsum disease gene". Nature Genetics. 17 (2): 185–9. doi:10.1038/ng1097-185. PMID 9326939.
- McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ (Dec 2010). "Structural studies on human 2-oxoglutarate dependent oxygenases". Current Opinion in Structural Biology. 20 (6): 659–72. doi:10.1016/j.sbi.2010.08.006. PMID 20888218.
Further reading
- Jansen GA, Mihalik SJ, Watkins PA, Jakobs C, Moser HW, Wanders RJ (Mar 1998). "Characterization of phytanoyl-Coenzyme A hydroxylase in human liver and activity measurements in patients with peroxisomal disorders". Clinica Chimica Acta; International Journal of Clinical Chemistry. 271 (2): 203–11. doi:10.1016/S0009-8981(97)00259-3. PMID 9565335.
- Jansen GA, Mihalik SJ, Watkins PA, Moser HW, Jakobs C, Denis S, Wanders RJ (Dec 1996). "Phytanoyl-CoA hydroxylase is present in human liver, located in peroxisomes, and deficient in Zellweger syndrome: direct, unequivocal evidence for the new, revised pathway of phytanic acid alpha-oxidation in humans". Biochemical and Biophysical Research Communications. 229 (1): 205–10. doi:10.1006/bbrc.1996.1781. PMID 8954107.
- Jansen GA, Ofman R, Ferdinandusse S, Ijlst L, Muijsers AO, Skjeldal OH, Stokke O, Jakobs C, Besley GT, Wraith JE, Wanders RJ (Oct 1997). "Refsum disease is caused by mutations in the phytanoyl-CoA hydroxylase gene". Nature Genetics. 17 (2): 190–3. doi:10.1038/ng1097-190. PMID 9326940.
- Mihalik SJ, Rainville AM, Watkins PA (Sep 1995). "Phytanic acid alpha-oxidation in rat liver peroxisomes. Production of alpha-hydroxyphytanoyl-CoA and formate is enhanced by dioxygenase cofactors". European Journal of Biochemistry / FEBS. 232 (2): 545–51. doi:10.1111/j.1432-1033.1995.545zz.x. PMID 7556205.