Peroxisome
A peroxisome (IPA: [pɛɜˈɹɒksɪˌsoʊm]) [1] is a membrane-bound organelle (formerly known as a microbody), found in the cytoplasm of virtually all eukaryotic cells.[2] Peroxisomes are oxidative organelles. Frequently, molecular oxygen serves as a co-substrate, from which hydrogen peroxide (H2O2) is then formed. Peroxisomes owe their name to hydrogen peroxide generating and scavenging activities. They perform key roles in lipid metabolism and the conversion of reactive oxygen species. Peroxisomes are involved in the catabolism of very long chain fatty acids, branched chain fatty acids, bile acid intermediates (in the liver), D-amino acids, and polyamines, the reduction of reactive oxygen species – specifically hydrogen peroxide.[3] – and the biosynthesis of plasmalogens, i.e., ether phospholipids critical for the normal function of mammalian brains and lungs [4] They also contain approximately 10% of the total activity of two enzymes (Glucose-6-phosphate dehydrogenase and 6-Phosphogluconate dehydrogenase) in the pentose phosphate pathway [5], which is important for energy metabolism.[4] It is vigorously debated whether peroxisomes are involved in isoprenoid and cholesterol synthesis in animals.[4] Other known peroxisomal functions include the glyoxylate cycle in germinating seeds ("glyoxysomes"), photorespiration in leaves,[6] glycolysis in trypanosomes ("glycosomes"), and methanol and/or amine oxidation and assimilation in some yeasts.
History
Peroxisomes (microbodies) were first described by a Swedish doctoral student, J. Rhodin in 1954.[7] They were identified as organelles by the Belgian cytologist Christian de Duve in 1967,[8] De Duve and co-workers discovered that peroxisomes contain several oxidases involved in the production of hydrogen peroxide (H2O2) as well as catalase involved in the decomposition of H2O2 to oxygen and water. Due to their role in peroxide metabolism, De Duve named them “peroxisomes”, replacing the formerly used morphological term “microbodies”. Later, it was described that firefly luciferase is targeted to peroxisomes in mammalian cells, allowing the discovery of the import targeting signal for peroxisomes, and triggering many advances in the peroxisome biogenesis field [9][10].
Structural Design
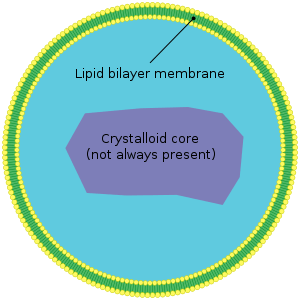
Peroxisomes are small (0.1-1 µm diameter) subcellular compartments (organelles) with a fine, granular matrix and surrounded by a single biomembrane which are located in the cytoplasm of a cell.[11][12] Compartmentalization creates an optimized environment to promote various metabolic reactions within peroxisomes required to sustain cellular functions and viability of the organism.
The number, size and protein composition of peroxisomes are variable and depend on cell type and environmental conditions. For example, in baker's yeast (S. cerevisiae), it has been observed that, with good glucose supply, only a few, small peroxisomes are present. In contrast, when the yeasts were supplied with long-chain fatty acids as sole carbon source up to 20 to 25 large peroxisomes can be formed.[13]
Metabolic functions
A major function of the peroxisome is the breakdown of very long chain fatty acids through beta oxidation. In animal cells, the long fatty acids are converted to medium chain fatty acids, which are subsequently shuttled to mitochondria where they eventually are broken down to carbon dioxide and water. In yeast and plant cells, this process is carried out exclusively in peroxisomes.[14][15]
The first reactions in the formation of plasmalogen in animal cells also occur in peroxisomes. Plasmalogen is the most abundant phospholipid in myelin. Deficiency of plasmalogens causes profound abnormalities in the myelination of nerve cells, which is one reason why many peroxisomal disorders affect the nervous system.[14] Peroxisomes also play a role in the production of bile acids important for the absorption of fats and fat-soluble vitamins, such as vitamins A and K. Skin disorders are features of genetic disorders affecting peroxisome function as a result [15].
The specific metabolic pathways that occur exclusively in mammalian peroxisomes are:[4]
- α-oxidation of phytanic acid
- β-oxidation of very long-chain and polyunsaturated fatty acids
- biosynthesis of plasmalogens
- conjugation of cholic acid as part of bile acid synthesis
Peroxisomes contain oxidative enzymes, such as D-amino acid oxidase and uric acid oxidase.[16] However the last enzyme is absent in humans, explaining the disease known as gout, caused by the accumulation of uric acid. Certain enzymes within the peroxisome, by using molecular oxygen, remove hydrogen atoms from specific organic substrates (labeled as R), in an oxidative reaction, producing hydrogen peroxide (H2O2, itself toxic):
Catalase, another peroxisomal enzyme, uses this H2O2 to oxidize other substrates, including phenols, formic acid, formaldehyde, and alcohol, by means of the peroxidation reaction:
- , thus eliminating the poisonous hydrogen peroxide in the process.
This reaction is important in liver and kidney cells, where the peroxisomes detoxify various toxic substances that enter the blood. About 25% of the ethanol that humans consume by drinking alcoholic beverages is oxidized to acetaldehyde in this way.[14] In addition, when excess H2O2 accumulates in the cell, catalase converts it to H2O through this reaction:
In higher plants, peroxisomes contain also a complex battery of antioxidative enzymes such as superoxide dismutase, the components of the ascorbate-glutathione cycle, and the NADP-dehydrogenases of the pentose-phosphate pathway. It has been demonstrated that peroxisomes generate superoxide (O2•−) and nitric oxide (•NO) radicals.[17][18]
There is evidence now that those reactive oxygen species including peroxisomal H2O2 are also important signalling molecules in plants and animals and contribute to healthy ageing and age-related disorders in humans.[19]
The peroxisome of plant cells is polarised when fighting fungal penetration. Infection causes a glucosinolate molecule to play an antifungal role to be made and delivered to the outside of the cell through the action of the peroxisomal proteins (PEN2 and PEN3).[20]
Peroxisomes in mammals and humans also contribute to anti-viral defense.[21] and the combat of pathogens [22]
Peroxisome assembly
Peroxisomes can be derived from the endoplasmic reticulum under certain experimental conditions and replicate by membrane growth and division out of pre-existing organelles.[23][24][25] Peroxisome matrix proteins are translated in the cytoplasm prior to import. Specific amino acid sequences (PTS or peroxisomal targeting signal) at the C-terminus (PTS1) or N-terminus (PTS2) of peroxisomal matrix proteins signals them to be imported into the organelle by a targeting factor. There are currently 36 known proteins involved in peroxisome biogenesis and maintenance, called peroxins [26], which participate in the process of peroxisome assembly in different organisms. In mammalian cells there are 13 characterized peroxins. In contrast to protein import into the endoplasmic reticulum (ER) or mitochondria, proteins do not need to be unfolded to be imported into the peroxisome lumen. The matrix protein import receptors, the peroxins PEX5 and PEX7, accompany their cargoes (containing a PTS1 or a PTS2 amino acid sequence, respectively) all the way to the peroxisome where they release the cargo into the peroxisomal matrix and then return to the cytosol – a step named recycling. A special way of peroxisomal protein targeting is called piggy backing. Proteins that are transported by this unique method do not have a canonical PTS, but rather bind on a PTS protein to be transported as a complex [27]. A model describing the import cycle is referred to as the extended shuttle mechanism.[28] There is now evidence that ATP hydrolysis is required for the recycling of receptors to the cytosol. Also, ubiquitination is crucial for the export of PEX5 from the peroxisome to the cytosol. The biogenesis of the peroxisomal membrane and the insertion of peroxisomal membrane proteins (PMPs) requires the peroxins PEX19, PEX3, and PEX16. PEX19 is a PMP receptor and chaperone, which binds the PMPs and routes them to the peroxisomal membrane, where it interacts with PEX3, a peroxisomal integral membrane protein. PMPs are then inserted into the peroxisomal membrane.
The degradation of peroxisomes is called pexophagy.[29]
Peroxisome interaction and communication
The diverse functions of peroxisomes require dynamic interactions and cooperation with many organelles involved in cellular lipid metabolism such as the endoplasmic reticulum (ER), mitochondria, lipid droplets, and lysosomes.[30]
Peroxisomes interact with mitochondria in several metabolic pathways, including β-oxidation of fatty acids and the metabolism of reactive oxygen species.[4] Both organelles are in close contact with the endoplasmic reticulum (ER) and share several proteins, including organelle fission factors.[31] Peroxisomes also interact with the endoplasmic reticulum (ER) and cooperate in the synthesis of ether lipids (plasmalogens) which are important for nerve cells (see above). In filamentous fungi, peroxisomes move on microtubules by 'hitchhiking,' a process involving contact with rapidly moving early endosomes. [32] Physical contact between organelles is often mediated by membrane contact sites, where membranes of two organelles are physically tethered to enable rapid transfer of small molecules, enable organelle communication and are crucial for coordination of cellular functions and hence human health.[33] Alterations of membrane contacts have been observed in various diseases.
Associated medical conditions
Peroxisomal disorders are a class of medical conditions that typically affect the human nervous system as well as many other organ systems. Two common examples are X-linked adrenoleukodystrophy and peroxisome biogenesis disorders.[34][35]
Genes
PEX genes encode the protein machinery ("peroxins") required for proper peroxisome assembly, as described above. Membrane assembly and maintenance requires three of these (peroxins 3, 16, and 19) and may occur without the import of the matrix (lumen) enzymes. Proliferation of the organelle is regulated by Pex11p.
Genes that encode peroxin proteins include: PEX1, PEX2 (PXMP3), PEX3, PEX5, PEX6, PEX7, PEX9 [36][37], PEX10, PEX11A, PEX11B, PEX11G, PEX12, PEX13, PEX14, PEX16, PEX19, PEX26, PEX28, PEX30, and PEX31. Between organisms, PEX numbering and function can differ.
Evolutionary origins
The protein content of peroxisomes varies across species or organism, but the presence of proteins common to many species has been used to suggest an endosymbiotic origin; that is, peroxisomes evolved from bacteria that invaded larger cells as parasites, and very gradually evolved a symbiotic relationship.[38] However, this view has been challenged by recent discoveries.[39] For example, peroxisome-less mutants can restore peroxisomes upon introduction of the wild-type gene.
Two independent evolutionary analyses of the peroxisomal proteome found homologies between the peroxisomal import machinery and the ERAD pathway in the endoplasmic reticulum [40][41], along with a number of metabolic enzymes that were likely recruited from the mitochondria.[41] Recently, it has been suggested that the peroxisome may have had an actinobacterial origin [42], however, this is controversial.[43]
Other related organelles
Other organelles of the microbody family related to peroxisomes include glyoxysomes of plants and filamentous fungi, glycosomes of kinetoplastids,[44] and Woronin bodies of filamentous fungi.
References
- "Definition of PEROXISOME". www.merriam-webster.com. Retrieved 2019-10-30.
- Islinger M, Voelkl A, Fahimi HD, Schrader M (November 2018). "The peroxisome: an update on mysteries 2.0". Histochemistry and Cell Biology. 150 (5): 443–471. doi:10.1007/s00418-018-1722-5. PMC 6182659. PMID 30219925.
- Bonekamp NA, Völkl A, Fahimi HD, Schrader M (2009). "Reactive oxygen species and peroxisomes: struggling for balance". BioFactors. 35 (4): 346–55. doi:10.1002/biof.48. PMID 19459143.
- Wanders RJ, Waterham HR (2006). "Biochemistry of mammalian peroxisomes revisited". Annual Review of Biochemistry. 75: 295–332. doi:10.1146/annurev.biochem.74.082803.133329. PMID 16756494.
- Antonenkov, Vasily D. (Jul 1989). "Dehydrogenases of the pentose phosphate pathway in rat liver peroxisomes". European Journal of Biochemistry. 183 (1): 75–82. doi:10.1111/j.1432-1033.1989.tb14898.x. ISSN 0014-2956. PMID 2753047.
- Evert RF, Eichhorn SE (2006). Esau's Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development. John Wiley & Sons. ISBN 9780471738435.
- Rhodin, J (1954). "Correlation of ultrastructural organization and function in normal and experimentally changed proximal tubule cells of the mouse kidney". Doctorate Thesis. Karolinska Institutet, Stockholm.
- de Duve C (April 1969). "The peroxisome: a new cytoplasmic organelle". Proceedings of the Royal Society of London. Series B, Biological Sciences. 173 (1030): 71–83. Bibcode:1969RSPSB.173...71D. doi:10.1098/rspb.1969.0039. PMID 4389648.
- Keller, G. A.; Gould, S.; Deluca, M.; Subramani, S. (May 1987). "Firefly luciferase is targeted to peroxisomes in mammalian cells". Proceedings of the National Academy of Sciences. 84 (10): 3264–3268. doi:10.1073/pnas.84.10.3264. ISSN 0027-8424. PMC 304849. PMID 3554235.
- Gould, S. J. (Sep 1988). "Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins". The Journal of Cell Biology. 107 (3): 897–905. doi:10.1083/jcb.107.3.897. ISSN 0021-9525. PMC 2115268. PMID 2901422.
- Karlson, P, Doenecke D, Koolman J, Fuchs G, Gerok W (2005). Karlsons Biochemistry and Pathobiochemistry (15 ed.). Stuttgart: Georg Thieme. pp. 396f. ISBN 978-3133578158. OCLC 181474420.
- Raven PH, Evert RF, Eichhorn SE (2006). Biology of Plants (4 ed.). Berlin: De Gruyter. pp. 53f. ISBN 978-3-11-018531-7. OCLC 180904366.
- Feldmann H (2009). Yeast: Molecular and Cell Biology. Weinheim: Wiley-VCH. p. 159. ISBN 978-3527326099. OCLC 489629727.
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "Chapter 12: Peroxisomes". Molecular Biology of the Cell (Fourth ed.). New York: Garland Science. ISBN 978-0-8153-3218-3.
- Schrader, Michael; Kamoshita, Maki; Islinger, Markus (Mar 2019). "Organelle interplay—peroxisome interactions in health and disease". Journal of Inherited Metabolic Disease. 0 (1): 71–89. doi:10.1002/jimd.12083. ISSN 1573-2665. PMC 7041636. PMID 30864148.
- del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ (November 1992). "Metabolism of oxygen radicals in peroxisomes and cellular implications". Free Radical Biology & Medicine. 13 (5): 557–80. doi:10.1016/0891-5849(92)90150-F. PMID 1334030.
- Corpas FJ, Barroso JB, del Río LA (April 2001). "Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells". Trends in Plant Science. 6 (4): 145–50. doi:10.1016/S1360-1385(01)01898-2. PMID 11286918.
- Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, et al. (September 2004). "Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants". Plant Physiology. 136 (1): 2722–33. doi:10.1104/pp.104.042812. PMC 523336. PMID 15347796.
- Lismont C, Revenco I, Fransen M (July 2019). "Peroxisomal Hydrogen Peroxide Metabolism and Signaling in Health and Disease". International Journal of Molecular Sciences. 20 (15): 3673. doi:10.3390/ijms20153673. PMC 6695606. PMID 31357514.
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, et al. (January 2009). "A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense". Science. 323 (5910): 101–6. Bibcode:2009Sci...323..101B. doi:10.1126/science.1163732. PMID 19095900.
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, et al. (May 2010). "Peroxisomes are signaling platforms for antiviral innate immunity". Cell. 141 (4): 668–81. doi:10.1016/j.cell.2010.04.018. PMC 3670185. PMID 20451243.
- Di Cara F, Bülow MH, Simmonds AJ, Rachubinski RA (November 2018). "Dysfunctional peroxisomes compromise gut structure and host defense by increased cell death and Tor-dependent autophagy". Molecular Biology of the Cell. 29 (22): 2766–2783. doi:10.1091/mbc.E18-07-0434. PMC 6249834. PMID 30188767.
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (July 2005). "Contribution of the endoplasmic reticulum to peroxisome formation". Cell. 122 (1): 85–95. doi:10.1016/j.cell.2005.04.025. PMID 16009135.
- Schrader M, Costello JL, Godinho LF, Azadi AS, Islinger M (May 2016). "Proliferation and fission of peroxisomes - An update". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1863 (5): 971–83. doi:10.1016/j.bbamcr.2015.09.024. PMID 26409486.
- Lazarow PB, Fujiki Y (Nov 1985). "Biogenesis of peroxisomes". Annual Review of Cell Biology. 1 (1): 489–530. doi:10.1146/annurev.cb.01.110185.002421. PMID 3916321.
- Saleem RA, Smith JJ, Aitchison JD (December 2006). "Proteomics of the peroxisome". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1763 (12): 1541–51. doi:10.1016/j.bbamcr.2006.09.005. PMC 1858641. PMID 17050007.
- Thoms, Sven (Nov 2015). "Import of proteins into peroxisomes: piggybacking to a new home away from home". Open Biology. 5 (11): 150148. doi:10.1098/rsob.150148. ISSN 2046-2441. PMC 4680570. PMID 26581572.
- Dammai V, Subramani S (April 2001). "The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol". Cell. 105 (2): 187–96. doi:10.1016/s0092-8674(01)00310-5. PMID 11336669.
- Eberhart T, Kovacs WJ (November 2018). "Pexophagy in yeast and mammals: an update on mysteries". Histochemistry and Cell Biology. 150 (5): 473–488. doi:10.1007/s00418-018-1724-3. hdl:20.500.11850/302080. PMID 30238155.
- Shai N, Schuldiner M, Zalckvar E (May 2016). "No peroxisome is an island - Peroxisome contact sites". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1863 (5): 1061–9. doi:10.1016/j.bbamcr.2015.09.016. PMC 4869879. PMID 26384874.
- Costello JL, Passmore JB, Islinger M, Schrader M (2018). "Multi-localized Proteins: The Peroxisome-Mitochondria Connection". Proteomics of Peroxisomes. Subcellular Biochemistry. 89. pp. 383–415. doi:10.1007/978-981-13-2233-4_17. ISBN 978-981-13-2232-7. PMID 30378033.
- Salogiannis J, Reck-Peterson SL (2017). "Hitchhiking: A Non-Canonical Mode of Microtubule-based Transport". Trends in Cell Biology. 27 (2): 141–150. doi:10.1016/j.tcb.2016.09.005. PMC 5258766. PMID 27665063.
- Castro IG, Schuldiner M, Zalckvar E (March 2018). "Mind the Organelle Gap - Peroxisome Contact Sites in Disease". Trends in Biochemical Sciences. 43 (3): 199–210. doi:10.1016/j.tibs.2018.01.001. PMC 6252078. PMID 29395653.
- Depreter M, Espeel M, Roels F (June 2003). "Human peroxisomal disorders". Microscopy Research and Technique. 61 (2): 203–23. doi:10.1002/jemt.10330. PMID 12740827.
- Islinger, Markus; Grille, Sandra; Fahimi, H. Dariush; Schrader, Michael (Mar 2012). "The peroxisome: an update on mysteries". Histochemistry and Cell Biology. 137 (5): 547–574. doi:10.1007/s00418-012-0941-4. hdl:10871/33969. ISSN 0948-6143. PMID 22415027.
- Effelsberg D, Cruz-Zaragoza LD, Schliebs W, Erdmann R (November 2016). "Pex9p is a new yeast peroxisomal import receptor for PTS1-containing proteins". Journal of Cell Science. 129 (21): 4057–4066. doi:10.1242/jcs.195271. PMID 27678487.
- Yifrach E, Chuartzman SG, Dahan N, Maskit S, Zada L, Weill U, et al. (November 2016). "Characterization of proteome dynamics during growth in oleate reveals a new peroxisome-targeting receptor". Journal of Cell Science. 129 (21): 4067–4075. doi:10.1242/jcs.195255. PMC 6275125. PMID 27663510.
- Lazarow PB, Fujiki Y (1985). "Biogenesis of peroxisomes". Annual Review of Cell Biology. 1: 489–530. doi:10.1146/annurev.cb.01.110185.002421. PMID 3916321.
- Fagarasanu A, Fagarasanu M, Rachubinski RA (2007). "Maintaining peroxisome populations: a story of division and inheritance". Annual Review of Cell and Developmental Biology. 23: 321–44. doi:10.1146/annurev.cellbio.23.090506.123456. PMID 17506702.
- Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A (April 2006). "The evolutionary origin of peroxisomes: an ER-peroxisome connection". Molecular Biology and Evolution. 23 (4): 838–45. doi:10.1093/molbev/msj103. PMID 16452116.
- Gabaldón T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (March 2006). "Origin and evolution of the peroxisomal proteome". Biology Direct. 1: 8. doi:10.1186/1745-6150-1-8. PMC 1472686. PMID 16556314.
- Duhita N, Le HA, Satoshi S, Kazuo H, Daisuke M, Takao S (January 2010). "The origin of peroxisomes: The possibility of an actinobacterial symbiosis". Gene. 450 (1–2): 18–24. doi:10.1016/j.gene.2009.09.014. PMID 19818387.
- Gabaldón T, Capella-Gutiérrez S (October 2010). "Lack of phylogenetic support for a supposed actinobacterial origin of peroxisomes". Gene. 465 (1–2): 61–5. doi:10.1016/j.gene.2010.06.004. PMID 20600706.
- Blattner J, Swinkels B, Dörsam H, Prospero T, Subramani S, Clayton C (December 1992). "Glycosome assembly in trypanosomes: variations in the acceptable degeneracy of a COOH-terminal microbody targeting signal". The Journal of Cell Biology. 119 (5): 1129–36. doi:10.1083/jcb.119.5.1129. PMC 2289717. PMID 1447292.
Further reading
- Innovative Training Network PERICO
- Schrader M, Costello J, Godinho LF, Islinger M (2015). "Peroxisome-mitochondria interplay and disease". J Inherit Metab Dis. 38 (4): 681–702. doi:10.1007/s10545-015-9819-7. hdl:10871/17472. PMID 25687155.
- Schrader M, Fahimi HD (2008). "The peroxisome: still a mysterious organelle". Histochem Cell Biol. 129 (4): 421–440. doi:10.1007/s00418-008-0396-9. PMC 2668598. PMID 18274771.
- Effelsberg D, Cruz-Zaragoza LD, Schliebs W, Erdmann R (2016). "Pex9p is a novel yeast peroxisomal import receptor for PTS1-proteins". Journal of Cell Science. 129 (21): 4057–4066. doi:10.1242/jcs.195271. PMID 27678487.
- Yifrach E, Chuartzman SG, Dahan N, Maskit S, Zada L, Weill U, Yofe I, Olender T, Schuldiner M, Zalckvar E (2016). "Characterization of proteome dynamics in oleate reveals a novel peroxisome targeting receptor". Journal of Cell Science. 129 (21): 4067–4075. doi:10.1242/jcs.195255. PMC 6275125. PMID 27663510.
- Mateos RM, León AM, Sandalio LM, Gómez M, del Río LA, Palma JM (December 2003). "Peroxisomes from pepper fruits (Capsicum annuum L.): purification, characterisation and antioxidant activity". Journal of Plant Physiology. 160 (12): 1507–16. doi:10.1078/0176-1617-01008. PMID 14717445.
- Corpas FJ, Barroso JB (2014). "Functional implications of peroxisomal nitric oxide (NO) in plants". Frontiers in Plant Science. 5: 97. doi:10.3389/fpls.2014.00097. PMC 3956114. PMID 24672535.
- Corpas FJ (November 2015). "What is the role of hydrogen peroxide in plant peroxisomes?". Plant Biology. 17 (6): 1099–103. doi:10.1111/plb.12376. PMID 26242708.

External links
| Wikiversity has learning resources about Peroxisomes at |
| Wikimedia Commons has media related to Peroxisomes. |