Plasmalogen
There are two types of ether phospholipids, plasmanyl- and plasmenyl-phospholipids. Plasmanyl-phospholipids have an ether bond in position sn-1 to an alkyl group. Plasmenyl-phospholipids have an ether bond in position sn-1 to an alkenyl group. The latter are called plasmalogens.[1][2][3]
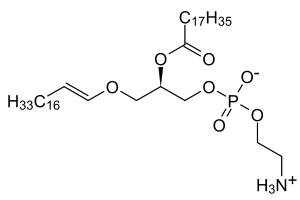
In mammals, the sn-1 position is typically derived from C16:0, C18:0, or C18:1 fatty alcohols while the sn-2 position is most commonly occupied by polyunsaturated fatty acids (PUFAs). The most common head groups present in mammalian plasmalogens are ethanolamine (designated plasmenylethalomines) or choline (designated plasmenylcholines).
Functions
Plasmalogens are found in numerous human tissues, with particular enrichment in the nervous, immune, and cardiovascular system.[1][2][3] In human heart tissue, nearly 30–40% of choline glycerophospholipids are plasmalogens. Even more striking is the fact that 32% of the glycerophospholipids in the adult human heart and 20% in brain and up to 70% of myelin sheath ethanolamine glycerophospholipids are plasmalogens.[4]
Although the functions of plasmalogens have not yet been fully elucidated, it has been demonstrated that they can protect mammalian cells against the damaging effects of reactive oxygen species.[1][2][3] In addition, they have been implicated as being signaling molecules and modulators of membrane dynamics.
History
Plasmalogens were first described by Feulgen and Voit in 1924 based on studies of tissue sections.[1] They treated these tissue sections with acid or mercuric chloride as part of a method to stain the nucleus. This resulted in the breakage of the plasmalogen vinyl-ether bond to yield aldehydes. In turn, the latter reacted with a fuchsine-sulfurous acid stain used in this nuclear staining method and gave rise to colored compounds inside the cytoplasm of the cells. Plasmalogens were named based on the fact that these colored compounds were present in the "plasmal" or inside of the cell.[1]
Biosynthesis
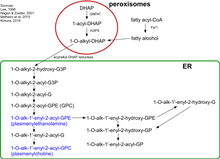
Biosynthesis of plasmalogens (PLs) begins with association of peroxisomal matrix enzymes GNPAT (glycerone phosphate acyl transferase) and AGPS (alkyl-glycerone phosphate synthase) on the luminal side of the peroxisomal membrane.[5] These two enzymes can physically interact with each other to increase efficiency. Therefore, fibroblasts without AGPS activity have a reduced GNPAT level and activity.[6][7]
The first step of the biosynthesis is catalyzed by GNPAT. This enzyme acylates dihydroxyacetone phosphate (DHAP) at the sn-1 position. This is followed by the exchange of the acyl group for an alkyl group by AGPS.[8] The 1-alkyl-DHAP is then reduced to 1-O-alkyl-2-hydroxy-sn-glycerophosphate (GPA) by an acyl/alkyl-DHAP reductase located in both peroxisomal and endoplasmatic reticulum (ER) membranes.[9] All other modifications occur in the ER. There an acyl group is placed at the sn-2 position by an alkyl/acyl GPA acyltransferase and the phosphate group is removed by a phosphatidic acid phosphatase to form 1-O-alkyl-2-acyl-sn-glycerol.
Using CDP-ethanolamine a phosphotransferase forms 1-O-alkyl-2-acyl-sn-GPEtn. After dehydrogenation at the 1- and 2-positions of the alkyl group by an electron transport system and plasmanylethanolamine desaturase the vinyl ether bond of plasmalogens is finally formed.The protein corresponding to plasmanylethanolamine desaturase has been identified and is called CarF in bacteria and TMEM189 in humans (and animals)[10]. Plasmenylcholine is formed from 1-O-alkyl-2-acyl-sn-glycerol by choline phosphotransferase. As there is no plasmenylcholine desaturase choline plasmalogens can be formed only after hydrolysis of ethanolamine PLs to 1-O-(1Z-alkenyl)-2-acyl-sn-glycerol that can be modified by choline phosphotransferase and CDP choline.[11][12]
Pathology
Peroxisome biogenesis disorders are autosomal recessive disorders often characterized by impaired plasmalogen biosynthesis. In these cases, the peroxisomal enzyme GNPAT, necessary for the initial steps of plasmalogen biosynthesis, is mislocalized to the cytoplasm where it is inactive. In addition, genetic mutations in the GNPAT or AGPS genes can result in plasmalogen deficiencies, which lead to the development of rhizomelic chondrodysplasia punctata (RCDP) type 2 or 3, respectively.[13] In such cases, both copies of the GNPAT or AGPS gene must be mutated in order for disease to manifest. Unlike the peroxisome biogenesis disorders, other aspects of peroxisome assembly in RCDP2 and RCDP3 patients are normal as is their ability to metabolize very long chain fatty acids. Individuals with severe plasmalogen deficiencies frequently show abnormal neurological development, skeletal malformation, impaired respiration, and cataracts. [14]
Deficits in plasmalogen levels contribute to pathology in Zellweger syndrome.[12]
Plasmalogen-knockout mice show similar alterations like arrest of spermatogenesis, development of cataract and defects in central nervous system myelination.[15][16]
During inflammation
During inflammation, neutrophil-derived myeloperoxidase produces hypochlorous acid (HOCl). HOCl causes oxidative chlorination of plasmalogens at the sn-1 chain by reacting with the vinyl ether bond.[17] Several researchers are currently investigating the impact of chlorinated lipids on pathology.
Possible disease links
The lack of good methods to assay plasmalogen has created difficulties for scientists to assess how plasmalogen might be involved in human diseases other than RCDP and Zellweger spectrum, in which the involvement is certain.[12] There is some evidence in humans that low plasmalogens are involved in the pathology of bronchopulmonary dysplasia, which is an important complication of premature birth.[12] and one study shows that plasmalogen levels are reduced in people with COPD who smoked compared with non-smokers. There is some evidence from humans and animals that there are reduced levels of plasmalogens in the brain in neurodegenerative disorders including Alzheimer disease, Parkinson's disease, Niemann–Pick disease, type C, Down syndrome, and multiple sclerosis, but as of 2012 it was not clear if the deficit was causing, or being caused by, the disease processes.[12]
Evolution
In addition to mammals, plasmalogens are also found in invertebrates and single cell organisms protozoans. Among bacteria they have been found in many anaerobic species including Clostridia, Megasphaera, and Veillonella. Among aerobic bacteria, plasmalogens occur in myxobacteria, and their plasmanylethanolamine desaturase (CarF) required to generate the vinyl ether bond, and hence plasmalogen, is conserved as TMEM189 in humans (and animals)[18]. Plasmalogens have been shown to have a complex evolutionary history based on the fact that their biosynthetic pathways differ in aerobic and anaerobic organisms.[19]
Recently, it has been demonstrated that the red blood cells of humans and great apes (chimpanzees, bonobos, gorillas and orangutans) have differences in their plasmalogen composition.[3] Total RBC plasmalogen levels were found to be lower in humans than in bonobos, chimpanzees, or gorillas, but higher than in orangutans. Gene expression data from all these species caused the authors to speculate that other human and great ape cells and tissues differ in plasmalogen levels. Although the consequences of these potential differences are unknown, cross-species differences in tissue plasmalogens could influence organ functions and multiple biological processes.
References
- Nagan, N.; Zoeller, R. A. (2001). "Plasmalogens: Biosynthesis and functions". Progress in Lipid Research. 40 (3): 199–229. doi:10.1016/S0163-7827(01)00003-0. PMID 11275267.
- Gorgas, K.; Teigler, A.; Komljenovic, D.; Just, W. W. (2006). "The ether lipid-deficient mouse: Tracking down plasmalogen functions". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1763 (12): 1511–1526. doi:10.1016/j.bbamcr.2006.08.038. PMID 17027098.
- Moser, A. B.; Steinberg, S. J.; Watkins, P. A.; Moser, H. W.; Ramaswamy, K.; Siegmund, K. D.; Lee, D. R.; Ely, J. J.; Ryder, O. A.; Hacia, J. G. (2011). "Human and great ape red blood cells differ in plasmalogen levels and composition". Lipids in Health and Disease. 10: 101. doi:10.1186/1476-511X-10-101. PMC 3129581. PMID 21679470.
- Farooqui, A. A.; Horrocks, L. A. (2001). "Plasmalogens: Workhorse lipids of membranes in normal and injured neurons and glia". The Neuroscientist : A Review Journal Bringing Neurobiology, Neurology and Psychiatry. 7 (3): 232–245. doi:10.1177/107385840100700308. PMID 11499402.
- P. Brites, H.R. Waterham, R.J. Wanders, Functions and biosynthesis of plasmalogens in health and disease, Biochim. Biophys. Acta 1636 (2004) 219–231.
- J. Biermann, W.W. Just, R.J. Wanders, H. Van Den Bosch, Alkyl-dihydroxyacetone phosphate synthase and dihydroxyacetone phosphate acyltransferase form a protein complex in peroxisomes, Eur. J. Biochem. 261 (1999) 492–499
- D. Hardeman, H. van den Bosch, Topography of ether phospholipid biosynthesis, Biochim. Biophys. Acta 1006 (1989) 1–8.
- A.J. Brown, F. Snyder, Alkyldihydroxyacetone-P synthase. Solubilization, partial purification, new assay method, and evidence for a ping-pong mechanism, J. Biol. Chem. 257 (1982) 8835–8839
- P.F. James, A.C. Lake, A.K. Hajra, L.K. Larkins, M. Robinson, F.G. Buchanan, R.A Zoeller, An animal cell mutant with a deficiency in acyl/alkyl-dihydroxyace- tone-phosphate reductase activity. Effects on the biosynthesis of ether-linked and diacyl glycerolipids, J. Biol. Chem. 272 (1997) 23540–23546
- Gallego-García A, Monera-Girona AJ, Pajares-Martínez E, Bastida-Martínez E, Pérez-Castaño R, Iniesta AA, Fontes M, Padmanabhan S, Elías-Arnanz M. "A bacterial light response reveals an orphan desaturase for human plasmalogen synthesis" (2019) Science 366 (6461) 128-132, doi:10.1126/science.aay1436, PMID=31604315
- T.C. Lee, Biosynthesis and possible biological functions of plasmalogens, Biochim. Biophys. Acta 1394 (1998) 129–145
- Braverman, NE; Moser, AB (September 2012). "Functions of plasmalogen lipids in health and disease". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1822 (9): 1442–52. doi:10.1016/j.bbadis.2012.05.008. PMID 22627108.
- Wanders, R.; Waterham, H. (2006). "Peroxisomal disorders: the single peroxisomal enzyme deficiencies". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1763 (12): 1707–20. doi:10.1016/j.bbamcr.2006.08.010. PMID 17055078.
- Rhizomelic Chondrodysplasia Punctata Type 1. Authors Braverman NE, Moser AB, Steinberg SJ. Editors In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. Source GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2014. 2001 Nov 16
- Gorgas K, Teigler A, Komljenovic D, Just WW. The ether lipid-deficient mouse: tracking down plasmalogen functions.Biochim Biophys Acta. 2006 Dec;1763(12):1511-26
- Rodemer C, Thai TP, Brugger B, Kaercher T, Werner H, Nave KA, Wieland F, Gorgas K, Just WW. Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice.Hum Mol Genet. 2003 Aug 1;12(15):1881-95.
- Albert, Carolyn J.; Crowley, Jan R.; Hsu, Fong-Fu; Thukkani, Arun K.; Ford, David A. (2001-06-29). "Reactive Chlorinating Species Produced by Myeloperoxidase Target the Vinyl Ether Bond of Plasmalogens IDENTIFICATION OF 2-CHLOROHEXADECANAL". Journal of Biological Chemistry. 276 (26): 23733–23741. doi:10.1074/jbc.M101447200. ISSN 0021-9258. PMID 11301330.
- Gallego-García A, Monera-Girona AJ, Pajares-Martínez E, Bastida-Martínez E, Pérez-Castaño R, Iniesta AA, Fontes M, Padmanabhan S, Elías-Arnanz M. "A bacterial light response reveals an orphan desaturase for human plasmalogen synthesis" (2019) Science 366 (6461) 128-132, doi:10.1126/science.aay1436, PMID=31604315
- Goldfine, H. (2010). "The appearance, disappearance and reappearance of plasmalogens in evolution". Progress in Lipid Research. 49 (4): 493–498. doi:10.1016/j.plipres.2010.07.003. PMID 20637230.
External links
- Plasmalogens at the US National Library of Medicine Medical Subject Headings (MeSH)