Phosphogluconate dehydrogenase
6-Phosphogluconate dehydrogenase (6PGD) is an enzyme in the pentose phosphate pathway. It forms ribulose 5-phosphate from 6-phosphogluconate:
- 6-phospho-D-gluconate + NAD(P)+ 6-phospho-2-dehydro-D-gluconate + NAD(P)H + H+
| 6PGD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
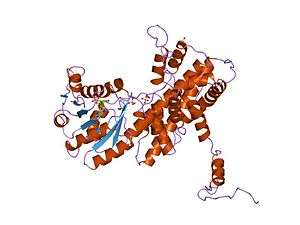 Crystallographic structure of sheep 6-phosphogluconate dehydrogenase complexed with adenosine 2'-monophosphate.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | 6PGD | ||||||||
| Pfam | PF00393 | ||||||||
| Pfam clan | CL0106 | ||||||||
| InterPro | IPR006114 | ||||||||
| PROSITE | PDOC00390 | ||||||||
| SCOPe | 2pgd / SUPFAM | ||||||||
| |||||||||
| Phosphogluconate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.1.1.44 | ||||||||
| CAS number | 9001-82-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| phosphogluconate dehydrogenase | |
|---|---|
| Identifiers | |
| Symbol | PGD |
| NCBI gene | 5226 |
| HGNC | 8891 |
| OMIM | 172200 |
| RefSeq | NM_002631 |
| UniProt | P52209 |
| Other data | |
| EC number | 1.1.1.44 |
| Locus | Chr. 1 p36.3-36.13 |
It is an oxidative carboxylase that catalyses the decarboxylating reduction of 6-phosphogluconate into ribulose 5-phosphate in the presence of NADP. This reaction is a component of the hexose mono-phosphate shunt and pentose phosphate pathways (PPP).[2][3] Prokaryotic and eukaryotic 6PGD are proteins of about 470 amino acids whose sequences are highly conserved.[4] The protein is a homodimer in which the monomers act independently:[3] each contains a large, mainly alpha-helical domain and a smaller beta-alpha-beta domain, containing a mixed parallel and anti-parallel 6-stranded beta sheet.[3] NADP is bound in a cleft in the small domain, the substrate binding in an adjacent pocket.[3]
Clinical significance
Mutations within the gene coding this enzyme result in 6-phosphogluconate dehydrogenase deficiency, an autosomal hereditary disease affecting the red blood cells.
As a possible drug target
6PGD is involved in cancer cell metabolism so 6PGD inhibitors have been sought.[5]
See also
- Parietin, a 6PGD inhibitor
References
- PDB: 1PGQ; Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C (July 1994). "Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism". Structure. 2 (7): 651–68. doi:10.1016/s0969-2126(00)00066-6. PMID 7922042.
- Broedel SE, Wolf RE (July 1990). "Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase". J. Bacteriol. 172 (7): 4023–31. doi:10.1128/jb.172.7.4023-4031.1990. PMC 213388. PMID 2113917.
- Adams MJ, Archibald IG, Bugg CE, Carne A, Gover S, Helliwell JR, Pickersgill RW, White SW (1983). "The three dimensional structure of sheep liver 6-phosphogluconate dehydrogenase at 2.6 A resolution". EMBO J. 2 (6): 1009–14. doi:10.1002/j.1460-2075.1983.tb01535.x. PMC 555222. PMID 6641716.
- Reizer A, Deutscher J, Saier MH, Reizer J (May 1991). "Analysis of the gluconate (gnt) operon of Bacillus subtilis". Mol. Microbiol. 5 (5): 1081–9. doi:10.1111/j.1365-2958.1991.tb01880.x. PMID 1659648.
- 6-Phosphogluconate dehydrogenase links oxidative PPP, lipogenesis and tumour growth by inhibiting LKB1–AMPK signalling. 2015
- Frampton EW, Wood WA (1961). "Carbohydrate oxidation by Pseudomonas fluorescens VI. Conversion of 2-keto-6-phosphogluconate to pyruvate". J. Biol. Chem. 236: 2571–2577. PMID 13894458.