Haplogroup K (mtDNA)
Haplogroup K is a human mitochondrial DNA (mtDNA) haplogroup. It is defined by the HVR1 mutations 16224C and 16311C.
| Haplogroup K | |
|---|---|
| Possible time of origin | 26,700 ± 4,300 years ago[1] |
| Possible place of origin | Possibly West Asia |
| Ancestor | U8b'K |
| Descendants | K1, K2 |
| Defining mutations | 3480 10550 11299 14798 16224 16311[2] |
Origin
Haplogroup K is believed to have originated in the mid-Upper Paleolithic, between about 30,000 and 22,000 years ago. It is the most common subclade of haplogroup U8b.[3] with an estimated age of c. 12,000 years BP.[4]
Distribution
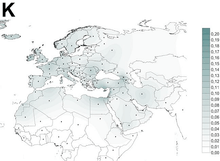
Haplogroup K appears in Central Europe, Southern Europe, Northern Europe, North Africa, the Horn of Africa, South Asia and West Asia and in populations with such an ancestry.
Haplogroup K is found in approximately 10% of native Europeans.[5][6]
Overall the mtDNA haplogroup K is found in about 6% of the population of Europe and the Near East, but it is more common in certain of these populations. Approximately 16% of the Druze of Syria, Lebanon, Israel, and Jordan, belong to haplogroup K.[7] It is also found among 8% of Palestinians.[8] Additionally, K reaches a level of 17% in Kurdistan.[9]
Approximately 32% of people with Ashkenazi Jewish ancestry are in haplogroup K. This high percentage points to a genetic bottleneck occurring some 100 generations ago.[7] Ashkenazi mtDNA K clusters into three subclades seldom found in non-Jews: K1a1b1a, K1a9, and K2a2a. Thus it is possible to detect three individual female ancestors, who were thought to be from a Hebrew/Levantine mtDNA pool, whose descendants lived in Europe.[10] A 2013 study however suggests these clades to instead originate from Western Europe.[11]
K appears to be highest in the Morbihan (17.5%) and Périgord-Limousin (15.3%) regions of France, and in Norway and Bulgaria (13.3%).[12] The level is 12.5% in Belgium, 11% in Georgia and 10% in Austria and Great Britain.[9]
Haplogroup K is also found among Gurage (10%),[8] Syrians (9.1%),[8] Afar (6.3%),[8] Zenata Berbers (4.11%),[13] Reguibate Sahrawi (3.70%),[13] Oromo (3.3%),[8] Iraqis (2.4%),[8] Saudis (0%-10.5%),[8] Yemenis (0%-9.8%),[8] and Algerians (0%-4.3%).[13]
Mtdna K was found in 0.9% of Beijing Han in a group of sampling.[14]
Ancient DNA
Haplogroup K has been found in the remains of three individuals from the Pre-Pottery Neolithic B site of Tell Ramad, Syria, dating from c. 6000 BC.[15] The clade was also discovered in skeletons of early farmers in Central Europe dated to around 5500-5300 BC, at percentages that were nearly double the percentage present in modern Europe. Some techniques of farming, together with associated plant and animal breeds, spread into Europe from the Near East. The evidence from ancient DNA suggests that the Neolithic culture spread by human migration.[16]
Analysis of the mtDNA of Ötzi, the frozen mummy from 3300 BC found on the Austrian-Italian border, has shown that Ötzi belongs to the K1 subclade. It cannot be categorized into any of the three modern branches of that subclade (K1a, K1b or K1c). The new subclade has provisionally been named K1ö for Ötzi.[17] Multiplex assay study was able to confirm that the Iceman's mtDNA belongs to a new European mtDNA clade with a very limited distribution amongst modern data sets.[18]
A woman buried some time between 2650 and 2450 BC in a presumed Amorite tomb at Terqa (Tell Ashara), Middle Euphrates Valley, Syria carried Haplogroup K.[19]
A lock of hair kept at a reliquary at Saint-Maximin-la-Sainte Baume basilica, France, which local tradition holds belonged to the biblical figure Mary Magdalene, was also assigned to haplogroup K. Ancient DNA sequencing of a capillary bulb bore the K1a1b1a subclade, indicating that she was likely of Pharisian maternal origin.[20]
Haplogroup K1 has likewise been observed among specimens at the mainland cemetery in Kulubnarti, Sudan, which date from the Early Christian period (AD 550-800).[21]
In 2016, researchers extracted the DNA from the tibia of two individuals separately dated to 7288-6771 BCE and 7605-7529 BCE buried in Theopetra cave, Greece, the oldest known human-made structure, and both individuals were found to belong to mtDNA Haplogroup K1c.[22]
Haplogroup K has also been observed among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the Pre-Ptolemaic/late New Kingdom and Roman periods.[23] Fossils excavated at the Late Neolithic site of Kelif el Boroud in Morocco, which have been dated to around 3,000 BCE, have likewise been observed to carry the K1 subclade.[24]
Subclades
Tree
This phylogenetic tree of haplogroup K subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[2] and subsequent published research.
| mtDNA HG "K" p-tree |
|---|
|
Genetic traits
A study involving Caucasian patients showed that individuals classified as haplogroup J or K demonstrated a significant decrease in risk of Parkinson's disease versus individuals carrying the most common haplogroup, H.[25]
In popular culture
In his popular book The Seven Daughters of Eve, Bryan Sykes named the originator of this mtDNA haplogroup Katrine.
On an 18 November 2005 broadcast of the Today Show, during an interview with Dr. Spencer Wells of The National Geographic Genographic Project, host Katie Couric was revealed to belong to haplogroup K.
On 14 August 2007, Stephen Colbert was told by geneticist Spencer Wells that he is a member of this haplogroup during a segment on The Colbert Report.
Henry Louis Gates Jr. states that Meryl Streep belongs to Haplogroup K in his book Faces of America.[26]
See also
| Wikimedia Commons has media related to Haplogroup K (mtDNA). |
- Genealogical DNA test
- Genetic genealogy
- Haplogroup K1a1b1a (mtDNA)
- Human mitochondrial genetics
- Population genetics
- Human mitochondrial DNA haplogroup
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- Behar et al. (2012), haplogroup.org
- van Oven, Mannis; Manfred Kayser (13 Oct 2008). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–E394. doi:10.1002/humu.20921. PMID 18853457. Archived from the original on 4 December 2012. Retrieved 2009-05-20.
- González, Ana M (2006). "The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country". BMC Genomics. 7: 124. doi:10.1186/1471-2164-7-124. PMC 1523212. PMID 16719915.
- Richards, M; Macaulay, V; Hickey, E; et al. (November 2000). "Tracing European founder lineages in the Near Eastern mtDNA pool". Am. J. Hum. Genet. 67: 1251–76. doi:10.1016/S0002-9297(07)62954-1. PMC 1288566. PMID 11032788.
- Bryan Sykes (2001). The Seven Daughters of Eve. London; New York: Bantam Press. ISBN 0393020185.
- "Maternal Ancestry". Oxford Ancestors. Archived from the original on 15 July 2017. Retrieved 7 February 2013.
- Skorecki, Karl; Quintana-Murci, Lluis; Pergola, Sergio Della; Kaplan, Matthew; Rosengarten, Dror; David Gurwitz; Richards, Martin; Bonne-Tamir, Batsheva; Villems, Richard; Garrigan, Daniel; Hammer, Michael F.; Behar, Doron M. (1 May 2004). "MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population". European Journal of Human Genetics. 12 (5): 355–364. doi:10.1038/sj.ejhg.5201156. PMID 14722586.
- Non, Amy. "ANALYSES OF GENETIC DATA WITHIN AN INTERDISCIPLINARY FRAMEWORK TO INVESTIGATE RECENT HUMAN EVOLUTIONARY HISTORY AND COMPLEX DISEASE" (PDF). University of Florida. Retrieved 17 April 2016.
- Lucia Simoni, Francesc Calafell, Davide Pettener, Jaume Bertranpetit, and Guido Barbujani, Geographic Patterns of mtDNA Diversity in Europe, American Journal of Human Genetics, vol. 66 (2000), pp. 262–278.
- Behar, DM; Metspalu, E; Kivisild, T; et al. (March 2006). "The matrilineal ancestry of Ashkenazi Jewry: portrait of a recent founder event". Am. J. Hum. Genet. 78 (3): 487–97. doi:10.1086/500307. PMC 1380291. PMID 16404693.
- Richards, Martin B.; Pereira, Luísa; Soares, Pedro; Carr, Martin; Macaulay, Vincent; Eng, Ken Khong; Woodward, Scott R.; Hatina, Jiři; Naumova, Oksana; Rychkov, Sergei; Perego, Ugo A.; Achilli, Alessandro; Olivieri, Anna; Fernandes, Verónica; Pala, Maria; Pereira, Joana B.; Costa, Marta D. (8 October 2013). "A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages". Nature Communications. 4: 2543. doi:10.1038/ncomms3543. PMC 3806353. PMID 24104924.
- Dubut, Vincent (2003). "mtDNA polymorphisms in five French groups: importance of regional sampling". European Journal of Human Genetics. 12 (4): 293–300. doi:10.1038/sj.ejhg.5201145. PMID 14694359.
- Asmahan Bekada; Lara R. Arauna; Tahria Deba; Francesc Calafell; Soraya Benhamamouch; David Comas (September 24, 2015). "Genetic Heterogeneity in Algerian Human Populations". PLoS ONE. 10 (9): e0138453. doi:10.1371/journal.pone.0138453. PMC 4581715. PMID 26402429.; S5 Table
- Rishishwar L, Jordan IK (2017). "Implications of human evolution and admixture for mitochondrial replacement therapy". BMC Genomics. 18 (1): 140. doi:10.1186/s12864-017-3539-3. PMC 5299762. PMID 28178941.
- Fernández Domínguez, Eva. "Polimorfismos de DNA mitocondrial en poblaciones antiguas de la cuenca mediterránea". Universitat de Barcelona. Retrieved 19 October 2017.
- W. Haak, et al, "Ancient DNA from the First European Farmers in 7500-Year-Old Neolithic Sites", Science, vol. 310, no. 5750 (2005), pp. 1016-1018; B. Bramanti, "Ancient DNA: Genetic analysis of aDNA from sixteen skeletons of the Vedrovice," Anthropologie, vol. 46, l no. 2-3 (2008), pp. 153-160; B. Bramanti et al, "Genetic Discontinuity Between Local Hunter-Gatherers and Central Europe’s First Farmers," Science, (published online 3 Sep 2009).
- Luca Ermini et al., "Complete Mitochondrial Genome Sequence of the Tyrolean Iceman," Current Biology, vol. 18, no. 21 (30 October 2008), pp. 1687-1693.
- Endicott et al., "Genotyping human ancient mtDNA control and coding region polymorphisms with a multiplexed Single-Base-Extension assay: the singular maternal history of the Tyrolean Iceman," BMC Genetics, vol. 10, no. 29 (19 June 2009).
- J. Tomczyk, et al., "Anthropological Analysis of the Osteological Material from an Ancient Tomb (Early Bronze Age) from the Middle Euphrates Valley, Terqa (Syria)," International Journal of Osteoarchaeology, published online ahead of print (2010).
- Lucotte, Gérard (December 2016). "The Mitochondrial DNA Mitotype of Sainte Marie-Madeleine" (PDF). International Journal of Sciences. 5 (12). Retrieved 16 February 2017.
- Sirak, Kendra; Frenandes, Daniel; Novak, Mario; Van Gerven, Dennis; Pinhasi, Ron (2016). Abstract Book of the IUAES Inter-Congress 2016 - A community divided? Revealing the community genome(s) of Medieval Kulubnarti using next- generation sequencing. IUAES.
- Hofmanová, Zuzana; Kreutzer, Susanne; Hellenthal, Garrett; Sell, Christian; Diekmann, Yoan; Díez-del-Molino, David; van Dorp, Lucy; López, Saioa; Kousathanas, Athanasios; Link, Vivian; Kirsanow, Karola; Cassidy, Lara M.; Martiniano, Rui; Strobel, Melanie; Scheu, Amelie; Kotsakis, Kostas; Halstead, Paul; Triantaphyllou, Sevi; Kyparissi-Apostolika, Nina; Urem-Kotsou, Dushka; Ziota, Christina; Adaktylou, Fotini; Gopalan, Shyamalika; Bobo, Dean M.; Winkelbach, Laura; Blöcher, Jens; Unterländer, Martina; Leuenberger, Christoph; Çilingiroğlu, Çiler; Horejs, Barbara; Gerritsen, Fokke; Shennan, Stephen J.; Bradley, Daniel G.; Currat, Mathias; Veeramah, Krishna R.; Wegmann, Daniel; Thomas, Mark G.; Papageorgopoulou, Christina; Burger, Joachim (2016). "Early farmers from across Europe directly descended from Neolithic Aegeans". Proceedings of the National Academy of Sciences. 113 (25): 6886–6891. doi:10.1073/pnas.1523951113. ISSN 0027-8424. PMC 4922144. PMID 27274049.
- Schuenemann, Verena J.; et al. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications. 8: 15694. doi:10.1038/ncomms15694. PMC 5459999. PMID 28556824.
- Fregel; et al. (2018). "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe" (PDF). bioRxiv 10.1101/191569.
- van der Walt, Joelle M.; Nicodemus, Kristin K.; Martin, Eden R.; Scott, William K.; Nance, Martha A.; Watts, Ray L.; Hubble, Jean P.; Haines, Jonathan L.; Koller, William C.; Lyons, Kelly; Pahwa, Rajesh; Stern, Matthew B.; Colcher, Amy; Hiner, Bradley C.; Jankovic, Joseph; Ondo, William G.; Allen Jr., Fred H.; Goetz, Christopher G.; Small, Gary W.; Mastaglia, Frank; Stajich, Jeffrey M.; McLaurin, Adam C.; Middleton, Lefkos T.; Scott, Burton L.; Schmechel, Donald E.; Pericak-Vance, Margaret A.; Vance, Jeffery M. (2003). "Mitochondrial Polymorphisms Significantly Reduce the Risk of Parkinson Disease". The American Journal of Human Genetics. 72 (4): 804–811. doi:10.1086/373937. ISSN 0002-9297. PMC 1180345. PMID 12618962.
- Gates, Henry Louis Jr. (2010). Faces of America: How 12 Extraordinary People Discovered their Pasts. NYU Press. p. 49. ISBN 978-0-8147-3265-6.CS1 maint: ref=harv (link)
- Behar DM, van Owen M, Rosset, Metspalu M, et al. (2012). "A "Copernican" reassessment of the human mitochondrial DNA tree from its root". Am J Hum Genet. 90 (4): 675–684. doi:10.1016/j.ajhg.2012.03.002. PMC 3322232. PMID 22482806.
- Soares, P; Ermini, L; Thomson, N; Mormina, M; Rito, T; Röhl, A; Salas, A; Oppenheimer, S; et al. (2009). "Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock". American Journal of Human Genetics. 84 (6): 740–59. doi:10.1016/j.ajhg.2009.05.001. PMC 2694979. PMID 19500773.
External links
- General
- Mannis van Oven's Phylotree
- Haplogroup K
- Spread of Haplogroup K, from National Geographic
- mtDNA Haplogroup K Project at Family Tree DNA
- Danish Demes Regional DNA Project: mtDNA Haplogroup K