Haplogroup U (mtDNA)
Haplogroup U is a human mitochondrial DNA haplogroup (mtDNA). The clade arose from haplogroup R, likely during the early Upper Paleolithic. Its various subclades (labelled U1–U9, diverging over the course of the Upper Paleolithic) are found widely distributed across Northern and Eastern Europe, Central, Western and South Asia, as well as North Africa, the Horn of Africa, and the Canary Islands.
| Haplogroup U | |
|---|---|
| Possible time of origin | 46,500 ± 3,300 years ago[1] |
| Possible place of origin | Western Asia[2] |
| Ancestor | R |
| Descendants | U1, U5, U6, U2'3'4'7'8'9 |
| Defining mutations | 11467, 12308, 12372[3] |
Origins
Haplogroup U descends from the haplogroup R mtDNA branch of the phylogenetic tree. The defining mutations (A11467G, A12308G, G12372A) are estimated to have arisen between 43,000 and 50,000 years ago, in the early Upper Paleolithic (around 46,530 ± 3,290 years before present, with a 95% confidence interval per Behar et al., 2012).
Ancient DNA classified as belonging to the U* mitochondrial haplogroup has been recovered from human skeletal remains found in Western Siberia, which have been dated to c. 45,000 years ago.[4] The mitogenome (33-fold coverage) of the Peştera Muierii 1 individual (PM1) from Romania (35 ky cal BP) has been identified as the basal haplogroup U6* not previously found in any ancient or present-day humans.[5]
Haplogroup U has been found among Iberomaurusian specimens dating from the Epipaleolithic at the Taforalt and Afalou prehistoric sites.[6] Among the Taforalt individuals, around 13% of the observed haplotypes belonged to various U subclades, including U4a2b (1/24; 4%), U4c1 (1/24; 4%), and U6d3 (1/24; 4%). A further 41% of the analysed haplotypes could be assigned to either haplogroup U or haplogroup H. Among the Afalou individuals, 44% of the analysed haplotypes could be assigned to either haplogroup U or haplogroup H (3/9; 33%).[7]
Haplogroup U has also been observed among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, dated to the 1st millennium BC.[8]
Additionally, haplogroup U has been observed in ancient Guanche fossils excavated in Gran Canaria and Tenerife on the Canary Islands, which have been radiocarbon-dated to between the 7th and 11th centuries CE. All of the clade-bearing individuals were inhumed at the Tenerife site, with these specimens found to belong to the U6b1a (4/7; 57%) and U6b (1/7; 14%) subclades.[9]
Distribution
Haplogroup U is found in 15% of Indian caste and 8% of Indian tribal populations.[10] Haplogroup U is found in approximately 11% of native Europeans and is held as the oldest maternal haplogroup found in that region.[10][11][12] In a 2013 study, all but one of the ancient modern human sequences from Europe belonged to maternal haplogroup U, thus confirming previous findings that haplogroup U was the dominant type of Mitochondrial DNA (mtDNA) in Europe before the spread of agriculture into Europe and the presence and the spread of the Indo-Europeans in Western Europe.[13][14]
Haplogroup U has various subclades numbered U1 to U9. Haplogroup K is a subclade of U8.[15] The old age has led to a wide distribution of the descendant subgroups across Western Eurasia, North Africa, and South Asia. Some subclades of haplogroup U have a more specific geographic range.
Subclades
Subclades are labelled U1–U9; Haplogroup K is a subclade of U8.
Van Oven and Kayser (2009) proposed subclades "U2'3'4'7'8" and "U4'9".[3] Behar et al. (2012) amended this by grouping "U4'9" as subordinate to "U2'3'4'7'8" for a new intermediate subclade "U2'3'4'7'8'9".
Haplogroup U1
The U1 subclades are: U1a (with deep-subclades U1a1, U1a1a, U1a1a1, U1a1b)[16] and U1b.[16]
Haplogroup U1 estimated to have arisen between 26,000 and 37,000 years ago. It is found at very low frequency throughout Europe. It is more often observed in eastern Europe, Anatolia and the Near East. It is also found at low frequencies in India. U1 is found in the Svanetia region of Georgia at 4.2%. Subclade U1a is found from India to Europe, but is extremely rare among the northern and Atlantic fringes of Europe including the British Isles and Scandinavia. Several examples in Tuscany have been noted. In India, U1a has been found in the Kerala region. U1b has a similar spread but is rarer than U1a. Some examples of U1b have been found among Jewish diaspora. Subclades U1a and U1b appear in equal frequency in eastern Europe.[17]
The rare U1 clade is also found among Algerians in Oran (0.83%-1.08%) and the Reguibat tribe of the Sahrawi (0.93%).[18]
The U1a1a subclade has been observed in an ancient individual excavated at the Kellis 2 cemetery in the Dakleh Oasis, located in the southwestern desert of Egypt. 21 of the Kellis burials have been radiocarbon-dated to around 80-445 AD, a timeframe within the Romano-Christian period.[19] Haplogroup U1 has also been found among specimens at the mainland cemetery in Kulubnarti, Sudan, which date from the Early Christian period (AD 550-800).[20]
DNA analysis of excavated remains now located at ruins of the Church of St. Augustine in Goa, India have also revealed the unique mtDNA subclade U1b. This sublineage is absent in India, but present in Georgia and surrounding regions.[21] Since the genetic analysis corroborates archaeological and literary evidence, it is believed that the excavated remains belong to Ketevan the Martyr, queen of Georgia.[21]
Haplogroup U5
The age of U5 is estimated at between 25,000 and 35,000 years old,[22] roughly corresponding to the Gravettian culture. Approximately 11% of Europeans (10% of European-Americans) have some variant of haplogroup U5.
U5 has been found in human remains dating from the Mesolithic in England, Germany, Lithuania, Poland, Portugal, Russia,[23] Sweden,[24] France[25] and Spain.[26] Neolithic skeletons (~7,000 years old) that were excavated from the Avellaner cave in Catalonia, northeastern Spain included a specimen carrying haplogroup U5.[27]
Haplogroup U5 and its subclades U5a and U5b today form the highest population concentrations in the far north, among Sami, Finns, and Estonians. However, it is spread widely at lower levels throughout Europe. This distribution, and the age of the haplogroup, indicate individuals belonging to this clade were part of the initial expansion tracking the retreat of ice sheets from Europe around 10,000 years ago.
The modern Basques and Cantabrians possess almost exclusively U5b lineages (U5b1f, U5b1c1, U5b2).[28][29]
Additionally, haplogroup U5 is found in small frequencies and at much lower diversity in the Near East and parts of northern Africa (areas with sizable U6 concentrations), suggesting back-migration of people from Europe toward the south.[30]
Mitochondrial haplogroup U5a has also been associated with HIV infected individuals displaying accelerated progression to AIDS and death.[31]
U5 was the main haplogroup of mesolithic European hunter gatherers. U haplogroups were present at 83% in European hunter gatherers before influx of Middle Eastern farmer and steppe Indo-European ancestry decreased its frequency to less than 21%.[19]
- U5 has polymorphisms in the locations of 3197 9477 13617 16192 16270
- U5a arose around 17,000 and 27,000 years ago and has polymorphisms in 14793 16256 ( + U5 polymorphisms).[32]
- U5a1 arose between 14,000 and 20,000 years ago and has polymorphisms in 15218 16399 ( + U5a polymorphisms).
- U5a1a arose between 8,000 and 16,000 years ago and has polymorphisms in 1700 16192 ( + U5a1 polymorphisms).
- U5a1a1 arose between 3,000 and 11,000 years ago and has polymorphisms in 5495 15924 ( + U5a1a polymorphisms).
- U5a1a1a arose less than 6,000 years ago[33] and has polymorphisms in 3816 (A3816G) (and has lost its polymorphism in 152 (backmutation) + U5a1a1 polymorphisms).
- U5a1a1b arose around between 600 and 6,000 years ago and has polymorphisms in 15110 (G15110A) (and has lost its polymorphism in 152 (backmutation) + U5a1a1 polymorphisms).
- U5a1a1c has polymorphisms in 6905 (A6905G) 13015 (T13015C) ( + U5a1a1 polymorphisms).
- U5a1a1d arose less than 4,300 years ago and has polymorphisms: G185A T204C T16362C (and has lost its polymorphism in 152 (backmutation) + U5a1a1 polymorphisms).
- U5a1a2 arose between 7,000 and 14,000 years ago and has polymorphisms in 573.1C (deletion) 12346 ( + U5a1a polymorphisms).
- U5a1a2a arose less than 5,400 years ago and has polymorphisms in 5319 6629 6719 ( + U5a1a2 polymorphisms).
- U5a1a2a1 arose less than 3,400 years ago and has polymorphisms T6293C ( + U5a1a2a polymorphisms).
- U5a1a2a arose less than 5,400 years ago and has polymorphisms in 5319 6629 6719 ( + U5a1a2 polymorphisms).
- U5a1a1 arose between 3,000 and 11,000 years ago and has polymorphisms in 5495 15924 ( + U5a1a polymorphisms).
- U5a1b arose between 6,000 and 11,000 years ago and has polymorphisms in 9667 (A9667G) ( + U5a1 polymorphisms).
- U5a1b1 arose between 5,000 and 9,000 years ago and has polymorphisms in 16291 (C16291T) ( + U5a1b polymorphisms).
- U5a1b1a arose between 2,500 and 7,500 and has polymorphisms T4553C ( + U5a1b1 polymorphisms).
- U5a1b1a1 less than 4,000 years ago and has polymorphisms C14574T ( + U5a1b1a polymorphisms).
- U5a1b1b arose less than 8,000 years ago and has polymorphisms in 8119 (T8119C) ( + U5a1b1 polymorphisms).
- U5a1b1c arose between 3,000 and 7,000 years ago and has polymorphisms in 9055 (G9055A) ( + U5a1b1 polymorphisms).
- U5a1b1c1 arose less than 5,000 years ago and has polymorphisms in 1187 (T1187C) ( + U5a1b1c polymorphisms).
- U5a1b1c2 arose less than 5,000 years ago and has polymorphisms in 3705 (G3705A) ( + U5a1b1c polymorphisms).
- U5a1b1d has polymorphisms in 12358 16093 ( + U5a1b1 polymorphisms).
- U5a1b1e has polymorphisms in 12582 16192 16294 ( + U5a1b1 polymorphisms).
- U5a1b1a arose between 2,500 and 7,500 and has polymorphisms T4553C ( + U5a1b1 polymorphisms).
- U5a1b2 has polymorphisms in 9632 ( + U5a1b polymorphisms).
- U5a1b3 has polymorphisms in 16362 16428 ( + U5a1b polymorphisms).
- U5a1b1 arose between 5,000 and 9,000 years ago and has polymorphisms in 16291 (C16291T) ( + U5a1b polymorphisms).
- U5a1c arose around 13000 years ago and has polymorphisms in 16320 ( + U5a1 polymorphisms).
- U5a1c1 has polymorphisms in 195 13802 ( + U5a1c polymorphisms).
- U5a1c2 has polymorphisms in 961 965.1C (deletion) ( + U5a1c polymorphisms).
- U5a1d arose around 19000 years ago and has polymorphisms in 3027 ( + U5a1 polymorphisms).
- U5a1d1 has polymorphisms in 5263 13002 (to adenosine) ( + U5a1d polymorphisms).
- U5a1d2 has polymorphisms in 573.1C (deletion) 3552 ( + U5a1d polymorphisms).
- U5a1d2a has polymorphisms in 195 4823 5583 16145 16189 ( + U5a1d2 polymorphisms).
- U5a1e has polymorphisms in 3564 8610 ( + U5a1 polymorphisms).
- U5a1f has polymorphisms in 6023 ( + U5a1 polymorphisms).
- U5a1a arose between 8,000 and 16,000 years ago and has polymorphisms in 1700 16192 ( + U5a1 polymorphisms).
- U5a2 arose around 14000 years ago and has polymorphisms in 16526 ( + U5a polymorphisms).
- U5a2a arose around 6000 years ago and has polymorphisms in 13827 13928C 16114 16294 ( + U5a2 polymorphisms). It has been found in an ancient Mesolithic sample (6000-5000 cal BCE) from the Cave of Santimamiñe in the Basque Country, Spain.[34]
- U5a2b arose around 8000 years ago and has polymorphisms in 9548 ( + U5a2 polymorphisms).
- U5a2c arose around 13000 years ago and has polymorphisms in 10619 ( + U5a2 polymorphisms).
- U5a2d and has polymorphisms in 7843 7978 8104 11107 16192! (backmutated in 16192 to the original Cambridge sequence) ( + U5a2 polymorphisms).
- U5a2e and has polymorphisms in 151 152 3768 15289 16189 16311 16362 ( + U5a2 polymorphisms).
- U5a1 arose between 14,000 and 20,000 years ago and has polymorphisms in 15218 16399 ( + U5a polymorphisms).
- U5b arose between 19,000 and 26,000 years ago[35] and has polymorphisms in 150 7768 14182 ( + U5 polymorphisms). Found among Siwa Berbers of the Siwa Oasis.[36]
- U5b1 arose between 11,000 and 20,000 years ago[37] and has polymorphisms in 5656 ( + U5b polymorphisms).
- U5b1a has polymorphisms in 5656 15097, 16189 and has lost its polymorphism in 7028 (backmutation) ( + U5b1 polymorphisms).
- U5b1b: has been found in Saami of Scandinavia, Finnish and the Berbers of North Africa, which were found to share an extremely young branch, aged merely ∼9,000 years. U5b1b was also found in Fulbe and Papel people in Guinea-Bissau and Yakuts people of northeastern Siberia.[38][39] It arose around 11000 years ago and has polymorphisms in 12618 16189 ( + U5b1 polymorphisms).
- U5b1c has polymorphisms in 5656 15191, 16189, 16311 ( + U5b1 polymorphisms) and arose about 13,000 years ago.
- U5b1d has polymorphisms in 5437 5656 and has lost its polymorphism in 16192 (backmutation) ( + U5b1 polymorphisms).
- U5b1e has polymorphisms in 152 2757 10283 12616 16189 and has lost its polymorphism in 16192 (backmutation) ( + U5b1 polymorphisms) and arose about 6600 years ago. U5b1e is mainly seen in central Europe among Czechs, Slovaks, Hungarians and southern Russians.[40]
- U5b1g has polymorphisms in 151 228 573.1C 5656 10654 13759 14577 ( + U5b1 polymorphisms).
- U5b2 arose between 17,000 and 23,000 years ago[41] and has polymorphisms in 1721 13637( + U5b polymorphisms). The clade has been found in remains dating from prehistoric times in Europe, such as the subclade U5b2c1 of La Braña man (found at the La Braña site in Spain). U5b2 is rare among French Basques (2.5%) and more frequent in the Spanish Basques.
- U5b2a between 12,000 and 19,000 years ago,[42] prevalent in Central Europe.[40]
- U5b2a1 between 9,000 and 18,000 years ago, descendants U5b2a1a, U5b2a1b.
- U5b2a2 between 7,000 and 14,000 years ago, descendants U5b2a2a, U5b2a2b, U5b2a2c. Frequent in Central Europe, U5b2a2a1 especially in Poland.[40]
- U5b2a3 between 3,000 and 14,000 years ago, descendant U5b2a3a.
- U5b2a4 between 1,000 and 10,000 years ago, descendant U5b2a4a.
- U5b2a5 les than 2,600 years ago, descendant U5b2a5a.
- U5b2a6 less than 12,000 years ago.
- U5b2b between 12,000 and 17,000 years ago.[43]
- U5b2c between 7,000 and 18,000 years ago.[44]
- U5b2c1 less than 8,000 years ago.[45] Found in a Phoenician individual from a Carthage tomb in Byrsa Hill, Tunisia.[46]
- U5b2a between 12,000 and 19,000 years ago,[42] prevalent in Central Europe.[40]
- U5b3: The subclade likely originates in the Italian peninsula,[40] and is mainly found among the Sardinians.[47]
- U5b1 arose between 11,000 and 20,000 years ago[37] and has polymorphisms in 5656 ( + U5b polymorphisms).
- U5a arose around 17,000 and 27,000 years ago and has polymorphisms in 14793 16256 ( + U5 polymorphisms).[32]
Haplogroup U6
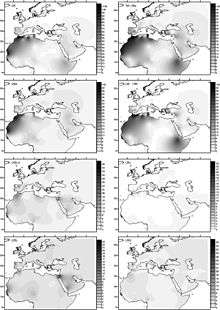
Haplogroup U6 was dated to between 31,000 and 43,000 years ago by Behar et al. (2012). Basal U6* was found in a Romanian specimen of ancient DNA (Peștera Muierilor) dated to 35,000 years ago.[48] Hervella et al. (2016) take this find as evidence for Paleolithic back-migration of Homo sapiens from Eurasia into Africa. The discovery of basal U6* in ancient DNA contributed to setting back the estimated age of U6 to around 46,000 years ago.[49]
Haplogroup U6 is common (with a prevalence of around 10%)[30] in Northwest Africa (with a maximum of 29% in an Algerian Mozabites[50]) and the Canary Islands (18% on average with a peak frequency of 50.1% in La Gomera). It is also found in the Iberian peninsula, where it has the highest diversity (10 out of 19 sublineages are only found in this region and not in Africa),[51] Northeast Africa and occasionally in other locations. U6 is also found at low frequencies in the Chad Basin, including the rare Canarian branch. This suggests that the ancient U6 clade bearers may have inhabited or passed through the Chad Basin on their way westward toward the Canary Islands.[52]
U6 is thought to have entered North Africa from the Near East around 30,000 years ago. It has been found among Iberomaurusian specimens dating from the Epipaleolithic at the Taforalt prehistoric site.[53] In spite of the highest diversity of Iberian U6, Maca-Meyer argues for a Near East origin of this clade based on the highest diversity of subclade U6a in that region,[51] where it would have arrived from West Asia, with the Iberian incidence primarily representing migration from the Maghreb and not persistence of a European root population.
According to Hernández et al. 2015 "the estimated entrance of the North African U6 lineages into Iberia at 10 ky correlates well with other L African clades, indicating that U6 and some L lineages moved together from Africa to Iberia in the Early Holocene."[54]
U6 has four main subclades:[51][53]
Subgroup U6a reflects the first African expansion from the Maghreb returning to the east. Derivative clade U6a1 signals a posterior movement from East Africa back to the Maghreb and the Near East. This migration coincides with the probable Afroasiatic linguistic expansion. U6b and U6c clades, restricted to West Africa, had more localized expansions. U6b probably reached the Iberian Peninsula during the Capsian diffusion in North Africa. Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.
— Maca-Meyer 2003
- U6a: subclade is the most widespread, stretching from the Canary Islands and Iberian Peninsula to the Horn of Africa and Near East. The subhaplogroup has its highest diversity in Northeast Africa. Ancient DNA analysis of Iberomaurusian skeletal remains at the Taforalt site in Morocco, which have been dated to the Later Stone Age between 15,100 and 13,900 ybp, observed the U6a subclade among most of the fossils (6/7; ~86%).[55] Fossils at the Early Neolithic site of Ifri n'Amr or Moussa in Morocco, which have been dated to around 5,000 BCE, have also been found to carry the U6a subhaplogroup. These ancient individuals bore an autochthonous Northwest African genomic component that peaks among modern Berbers, indicating that they were ancestral to populations in the area.[56] U6a's estimated age is 24-27,500 BP. It has one major subclade:
- U6b: shows a more patched and western distribution. In the Iberian peninsula, U6b is more frequent in the north, whereas U6a is more common in the south. It has also been found at low frequencies in Morocco, Algeria, Senegal and Nigeria. Estimated age: 8,500-24,500 BP. It has one subclade:
- U6b1: found only in the Canary Islands and in the Iberian peninsula. Estimated age: c. 6000 BP.
- U6c: only found in Morocco and Canary Islands. Estimated age: 6,000-17,500 BP.
- U6d: most closely related to U6b. Localized in the Maghreb, with a presence in Europe. It arose between 10,000 and 13,000 BP.
U6a, U6b and U6d share a common basal mutation (16219) that is not present in U6c, whereas U6c has 11 unique mutations. U6b and U6d share a mutation (16311) not shared by U6a, which has three unique mutations.
U2'3'4'7'8'9
Subclades U2, U3, U4, U7, U8 and U9 are now thought to be monophyletic, its common ancestor "U2'3'4'7'8'9" defined by mutation A1811G, arising between about 42,000 and 48,000 years ago (Behar et al., 2012). Within U2'3'4'7'8'9, U4 and U9 may be monophyletic, as "U4'9" (mutations T195C!, G499A, T5999C) arising between 31,000 and 43,000 years ago (Behar et al., 2012).
Haplogroup U2
Haplogroup U2 is most common in South Asia[58] but is also found in low frequency in Central and West Asia, as well as in Europe as U2e (the European variety of U2 is named U2e).[59] The overall frequency of U2 in South Asia is largely accounted for by the group U2i in India whereas haplogroup U2e, common in Europe, is rare; given that these lineages diverged approximately 50,000-years-ago, these data have been interpreted as indicating very low maternal-line gene-flow between South Asia and Europe throughout this period.[58] Approximately one half of the U mtDNAs in India belong to the Indian-specific branches of haplogroup U2 (U2i: U2a, U2b and U2c).[58] While U2 is typically found in India, it is also present in the Nogais, descendants of various Mongolic and Turkic tribes, who formed the Nogai Horde.[60] Both U2 and U4 are found in the Ket and Nganasan peoples, the indigenous inhabitants of the Yenisei River basin and the Taymyr Peninsula.[61]
The U2 subclades are: U2a,[62] U2b,[63] U2c,[64] U2d,[65] and U2e.[66] With the India-specific subclades U2a, U2b, and U2c collectively referred to as U2i, the Eurasian haplogroup U2d appears to be a sister clade with the Indian haplogroup U2c,[67] while U2e is considered a European-specific subclade but also found in South India.[59]
Haplogroup U2 has been found in the remains of a 37,000[68] and 30,000-year-old hunter-gatherer from the Kostyonki, Voronezh Oblast in Central-South European Russia., [69] in the remains of a 32,000-year-old hunter-gatherer from the mouth of river Yana,[70] in 4800 to 4000-year-old human remains from a Beaker culture site of the Late Neolithic in Kromsdorf Germany,[71] and in 2,000-year-old human remains from Bøgebjerggård in Southern Denmark. However, haplogroup U2 is rare in present-day Scandinavians.[72] The remains of a 2,000-year-old West Eurasian male of haplogroup U2e1 was found in the Xiongnu Cemetery of Northeast Mongolia.[73]
Haplogroup U3
Haplogroup U3 falls into two subclades:: U3a[74] and U3b.[74]
Coalescence age for U3a is estimated as 18,000 to 26,000-years-ago while the coalescence age for U3b is estimated as 18,000 to 24,000-years-ago. U3a is found in Europe, the Near East, the Caucasus and North Africa. The almost-entirely European distributed subclade, U3a1, dated at 4000 to 7000-years-ago, suggests a relatively recent (late Holocene or later) expansion of these lineages in Europe. There is a minor U3c subclade (derived from U3a), represented by a single Azeri mtDNA from the Caucasus. U3b is widespread across the Middle East and the Caucasus, and it is found especially in Iran, Iraq and Yemen, with a minor European subclade, U3b1b, dated at 2000 to 3000-years-ago.[75] Haplogroup U3 is defined by the HVR1 transition A16343G. It is found at low levels throughout Europe (about 1% of the population), the Near East (about 2.5% of the population), and Central Asia (about 1% of the population). U3 is present in the Svan population from the Svaneti region (about 4.2% of the population) and among Lithuanian Romani, Polish Romani, and Spanish Romani populations (36-56%)[76][77][32] consistent with a common migration route from India then out-of-the Balkans for the Lithuanian, Polish, and Spanish Roma.[78]
The U3 clade is also found among Mozabite Berbers (10.59%),[18] as well as Egyptians in the El-Hayez (2.9%)[79] and Gurna oases (2.9%),[80] and Algerians in Oran (1.08%-1.25%).[18] The rare U3a subclade occurs among the Tuareg inhabiting Niger (3.23%).[81]
Haplogroup U3 has been found in some of the 6400-year-old remains (U3a) discovered in the caves at Wadi El‐Makkukh near Jericho associated with the Chalcolithic period.[82] Haplogroup U3 was already present in the West Eurasian gene pool around 6,000-years-ago and probably also its subclade U3a as well.[82]
Haplogroup U4
Haplogroup U4 has its origin between 21,000 and 14,000 years ago. Its distribution is associated with the population bottleneck due to the Last Glacial Maximum.[76]
U4 has been found in ancient DNA,[83] and it is relatively rare in modern populations,[40] although it is found in substantial ratios in certain indigenous populations of Northern Asia and Northern Europe, being associated with the remnants of ancient European hunting-gatherers preserved in the indigenous populations of Siberia.[84][85][86] U4 is found in the Nganasan people of the Taymyr Peninsula,[61][87] in the Mansi (16.3%) an endangered people,[86] and in the Ket people (28.9%) of the Yenisei River.[86] It is found in Europe with highest concentrations in Scandinavia and the Baltic states.[88] and is found in the Sami population of the Scandinavian peninsula (although, U5b has a higher representation).[89] U4 is also preserved in the Kalash people (current population size 3,700)[90] a unique tribe among the Indo-Aryan peoples of Pakistan where U4 (subclade U4a1[91]) attains its highest frequency of 34%.[77][92]
The U4 subclades are: U4a,[93] U4b,[94] U4c,[95] and U4d.[96]
Haplogroup U4 is associated with ancient European hunter-gatherers and has been found in 7,200 to 6,000-year-old remains of the Pitted Ware culture in Gotland Sweden and in 4,400 to 3,800-year-old remains from the Damsbo site of the Danish Beaker culture.[97][24][98] Remains identified as subclade U4a2 are associated with the Corded Ware culture, which flourished 5200 to 4300 years ago in Eastern and Central Europe and encompassed most of continental northern Europe from the Volga River in the east to the Rhine in the west.[99] Mitochondrial DNA recovered from 3,500 to 3,300-year-old remains at the Bredtoftegård site in Denmark associated with the Nordic Bronze Age include haplogroup U4 with 16179T in its HVR1 indicative of subclade U4c1.[98][100][98][101]
Haplogroup U7
Haplogroup U7 is considered a West Eurasian-specific mtDNA haplogroup, believed to have originated in the Black Sea area approximately 30,000-years-ago.[58][102][103] In modern populations, U7 occurs at low frequency in the Caucasus,[103] the western Siberian tribes,[104] West Asia (about 4% in the Near East, while peaking with 10% in Iranians),[58] South Asia (about 12% in Gujarat, the westernmost state of India, while for the whole of India its frequency stays around 2%, and 5% in Pakistan),[58] and the Vedda people of Sri Lanka where it reaches it highest frequency of 13.33% (subclade U7a).[105] One third of the West Eurasian-specific mtDNAs found in India are in haplogroups U7, R2 and W. It is speculated that large-scale immigration carried these mitochondrial haplogroups into India.[58]
The U7 subclades are: U7a (with deep-subclades U7a1, U7a2, U7a2a, U7a2b)[106] and U7b.[106]
Genetic analysis of individuals associated with the Late Hallstatt culture from Baden-Württemberg Germany considered to be examples of Iron Age "princely burials" included haplogroup U7.[107] Haplogroup U7 was reported to have been found in 1200-year-old human remains (dating to around 834), in a woman believed to be from a royal clan who was buried with the Viking Oseberg Ship in Norway.[108] Haplogroup U7 was found in 1000-year-old human remains (dating to around AD 1000-1250) in a Christian cemetery is Kongemarken Denmark. However, U7 is rare among present-day ethnic Scandinavians.[104]
The U7a subclade is especially common among Saudis, constituting around 30% of maternal lineages in the Eastern Province.[109]
Haplogroup U8
- Haplogroup U8a: The Basques have the most ancestral phylogeny in Europe for the mitochondrial haplogroup U8a. This is a rare subgroup of U8, placing the Basque origin of this lineage in the Upper Palaeolithic. The lack of U8a lineages in Africa suggests that their ancestors may have originated from West Asia.[15]
- Haplogroup U8b: This clade has been found in Italy and Jordan.[15]
- Haplogroup U8b'K: This clade may be synonymous with Haplogroup K and Haplogroup UK.
The haplogroup U8b's most common subclade is haplogroup K, which is estimated to date to between 30,000 and 22,000 years ago. Haplogroup K makes up a sizeable fraction of European and West Asian mtDNA lineages. It is now known it is actually a subclade of haplogroup U8b'K,[15] and is believed to have first arisen in northeastern Italy. Haplogroup UK shows some evidence of being highly protective against AIDS progression.[31]
Haplogroup U9
Haplogroup U9 is a rare clade in mtDNA phylogeny, characterized only recently in a few populations of Pakistan (Quintana-Murci et al. 2004). Its presence in Ethiopia and Yemen, together with some Indian-specific M lineages in the Yemeni sample, points to gene flow along the coast of the Arabian Sea. Haplogroups U9 and U4 share two common mutations at the root of their phylogeny. It is interesting that, in Pakistan, U9 occurs frequently only among the so-called Makrani population. In this particular population, lineages specific to parts of Eastern Africa occur as frequently as 39%, which suggests that U9 lineages in Pakistan may have an African origin (Quintana-Murci et al. 2004). Regardless of which coast of the Arabian Sea may have been the origin of U9, its Ethiopian–southern Arabian–Indus Basin distribution hints that the subclade's diversification from U4 may have occurred in regions far away from the current area of the highest diversity and frequency of haplogroup U4—East Europe and western Siberia.[110]
See also
| Wikimedia Commons has media related to Haplogroup U (mtDNA). |
- Genealogical DNA test
- Genetic genealogy
- Human mitochondrial genetics
- Population genetics
- The Seven Daughters of Eve
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- Behar et al. (2012), note the revised estimate of 42–58 kya based on ancient DNA by Hervella et al. (2016).
- Hervella et al. (2016) "Individuals carrying haplogroup U possibly spread westward from Western Asia around 39–52 ky, reaching Europe as signaled by haplogroup U5, and North Africa signaled by haplogroup U6"
- van Oven M, Kayser M (February 2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386-94. doi:10.1002/humu.20921. PMID 18853457.
- Larruga JM, Marrero P, Abu-Amero KK, Golubenko MV, Cabrera VM (May 2017). "Carriers of mitochondrial DNA macrohaplogroup R colonized Eurasia and Australasia from a southeast Asia core area". BMC Evolutionary Biology. 17 (1): 115. doi:10.1186/s12862-017-0964-5. PMC 5442693. PMID 28535779. Marrero P, Abu-Amero KK, Larruga JM, Cabrera VM (November 2016). "Carriers of human mitochondrial DNA macrohaplogroup M colonized India from southeastern Asia". BMC Evolutionary Biology. 16 (1): 246. doi:10.1186/s12862-016-0816-8. PMC 5105315. PMID 27832758.
- Hervella M, Svensson EM, Alberdi A, Günther T, Izagirre N, Munters AR, et al. (May 2016). "The mitogenome of a 35,000-year-old Homo sapiens from Europe supports a Palaeolithic back-migration to Africa". Scientific Reports. 6: 25501. Bibcode:2016NatSR...625501H. doi:10.1038/srep25501. PMC 4872530. PMID 27195518..
- Secher B, Fregel R, Larruga JM, Cabrera VM, Endicott P, Pestano JJ, González AM (May 2014). "The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents". BMC Evolutionary Biology. 14: 109. doi:10.1186/1471-2148-14-109. PMC 4062890. PMID 24885141.
- Kefi R, Hechmi M, Naouali C, Jmel H, Hsouna S, Bouzaid E, et al. (January 2018). "On the origin of Iberomaurusians: new data based on ancient mitochondrial DNA and phylogenetic analysis of Afalou and Taforalt populations". Mitochondrial DNA. Part A, DNA Mapping, Sequencing, and Analysis. 29 (1): 147–157. doi:10.1080/24701394.2016.1258406. PMID 28034339.
- Schuenemann VJ, Peltzer A, Welte B, van Pelt WP, Molak M, Wang CC, et al. (May 2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications. 8: 15694. Bibcode:2017NatCo...815694S. doi:10.1038/ncomms15694. PMC 5459999. PMID 28556824.
- Rodríguez-Varela R, Günther T, Krzewińska M, Storå J, Gillingwater TH, MacCallum M, et al. (November 2017). "Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans". Current Biology. 27 (21): 3396–3402.e5. doi:10.1016/j.cub.2017.09.059. PMID 29107554.
- Karmin M (2005). Human mitochondrial DNA haplogroup R in India (PDF) (M.Sc. thesis). University of Tartu.
- Sykes B (2001). The Seven Daughters of Eve. London; New York: Bantam Press. ISBN 978-0393020182.
- "Maternal Ancestry". Oxford Ancestors. Archived from the original on 15 July 2017. Retrieved 7 February 2013.
- Fu Q, Mittnik A, Johnson PL, Bos K, Lari M, Bollongino R, et al. (April 2013). "A revised timescale for human evolution based on ancient mitochondrial genomes". Current Biology. 23 (7): 553–559. doi:10.1016/j.cub.2013.02.044. PMC 5036973. PMID 23523248.
- "Yamnaya, Light Skinned, Brown Eyed….Ancestors???". 2015-06-15.
- González AM, García O, Larruga JM, Cabrera VM (May 2006). "The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country". BMC Genomics. 7 (1): 124. doi:10.1186/1471-2164-7-124. PMC 1523212. PMID 16719915.
- Scott R (2010). "Phylogeny of mt-hg U3" (PDF).
- "mtDNA haplogroup U1 project". FamilyTreeDNA.
- Bekada A, Arauna LR, Deba T, Calafell F, Benhamamouch S, Comas D (September 24, 2015). "Genetic Heterogeneity in Algerian Human Populations". PLOS ONE. 10 (9): e0138453. Bibcode:2015PLoSO..1038453B. doi:10.1371/journal.pone.0138453. PMC 4581715. PMID 26402429.
- Molto JE, Loreille O, Mallott EK, Malhi RS, Fast S, Daniels-Higginbotham J, et al. (October 2017). "Complete Mitochondrial Genome Sequencing of a Burial from a Romano-Christian Cemetery in the Dakhleh Oasis, Egypt: Preliminary Indications". Genes. 8 (10): 262. doi:10.3390/genes8100262. PMC 5664112. PMID 28984839.
- Sirak K, Frenandes D, Novak M, Van Gerven D, Pinhasi R (2016). Abstract Book of the IUAES Inter-Congress 2016 - A community divided? Revealing the community genome(s) of Medieval Kulubnarti using next- generation sequencing. IUAES.
- Rai N, Taher N, Singh M, Chaubey G, Jha AN, Singh L, Thangaraj K (January 2014). "Relic excavated in western India is probably of Georgian Queen Ketevan". Mitochondrion. 14 (1): 1–6. doi:10.1016/j.mito.2013.12.002. PMID 24355295.
- "Haplogroup U5". haplogroup.org. 2016-06-06.
30,248.3 ± 5,330.5; CI=95% (Behar et al., 2012)
- Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, et al. (October 2009). "Genetic discontinuity between local hunter-gatherers and central Europe's first farmers". Science. 326 (5949): 137–40. Bibcode:2009Sci...326..137B. doi:10.1126/science.1176869. PMID 19729620.
- Malmström H, Gilbert MT, Thomas MG, Brandström M, Storå J, Molnar P, et al. (November 2009). "Ancient DNA reveals lack of continuity between neolithic hunter-gatherers and contemporary Scandinavians" (PDF). Current Biology. 19 (20): 1758–62. doi:10.1016/j.cub.2009.09.017. PMID 19781941.
- Deguilloux MF, Soler L, Pemonge MH, Scarre C, Joussaume R, Laporte L (January 2011). "News from the west: ancient DNA from a French megalithic burial chamber". American Journal of Physical Anthropology. 144 (1): 108–18. doi:10.1002/ajpa.21376. PMID 20717990.
- Sánchez-Quinto F, Schroeder H, Ramirez O, Avila-Arcos MC, Pybus M, Olalde I, et al. (August 2012). "Genomic affinities of two 7,000-year-old Iberian hunter-gatherers" (PDF). Current Biology. 22 (16): 1494–9. doi:10.1016/j.cub.2012.06.005. PMID 22748318.
- Lacan M, Keyser C, Ricaut FX, Brucato N, Tarrús J, Bosch A, et al. (November 2011). "Ancient DNA suggests the leading role played by men in the Neolithic dissemination". Proceedings of the National Academy of Sciences of the United States of America. 108 (45): 18255–9. Bibcode:2011PNAS..10818255L. doi:10.1073/pnas.1113061108. PMC 3215063. PMID 22042855.
- "Haplogroup U5". Eupedia.
- "Basque Genetics: Abstracts and Summaries". Kazharia.
- "The Genographic Project". National Geographic.
- Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, Poole JC, Lautenberger J, Sezgin E, et al. (November 2008). "Mitochondrial DNA haplogroups influence AIDS progression". AIDS. 22 (18): 2429–39. doi:10.1097/QAD.0b013e32831940bb. PMC 2699618. PMID 19005266.
- Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Miścicka-Sliwka D (March 2006). "Mitochondrial DNA diversity in the Polish Roma". Annals of Human Genetics. 70 (Pt 2): 195–206. doi:10.1111/j.1529-8817.2005.00222.x. PMID 16626330.
- Age: 1,398.1 ± 1,872.3; CI=95% (Behar et al., 2012) haplogroup.org
- Palencia-Madrid L, Cardoso S, Keyser C, López-Quintana JC, Guenaga-Lizasu A, de Pancorbo MM (May 2017). "Ancient mitochondrial lineages support the prehistoric maternal root of Basques in Northern Iberian Peninsula". European Journal of Human Genetics. 25 (5): 631–636. doi:10.1038/ejhg.2017.24. PMC 5437903. PMID 28272540.
- 22,794.0 ± 3,590.3; CI=95% (Behar et al., 2012) haplogroup.org
- Coudray C, Olivieri A, Achilli A, Pala M, Melhaoui M, Cherkaoui M, et al. (March 2009). "The complex and diversified mitochondrial gene pool of Berber populations". Annals of Human Genetics. 73 (2): 196–214. doi:10.1111/j.1469-1809.2008.00493.x. PMID 19053990.
- Age: 15,529.7 ± 4,889.9; CI=95% "mtDNA Haplogroup U5b1". haplogroup.org. 2016-06-06.
- Rosa A, Ornelas C, Jobling MA, Brehm A, Villems R (July 2007). "Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective". BMC Evolutionary Biology. 7 (1): 124. doi:10.1186/1471-2148-7-124. PMC 1976131. PMID 17662131.
- Achilli A, Rengo C, Battaglia V, Pala M, Olivieri A, Fornarino S, et al. (May 2005). "Saami and Berbers--an unexpected mitochondrial DNA link". American Journal of Human Genetics. 76 (5): 883–6. doi:10.1086/430073. PMC 1199377. PMID 15791543.
- Malyarchuk B, Derenko M, Grzybowski T, Perkova M, Rogalla U, Vanecek T, et al. (April 2010). Gilbert (ed.). "The peopling of Europe from the mitochondrial haplogroup U5 perspective". PLOS ONE. 5 (4): e10285. Bibcode:2010PLoSO...510285M. doi:10.1371/journal.pone.0010285. PMC 2858207. PMID 20422015.
Subcluster U5b2a is characterized by a predominantly central European distribution, since a large number of U5b samples from Poland, Slovakia and the Czech Republic fall into this subcluster. For instance, subcluster U5b2a2 is frequent in central Europe (with the highest frequency of its subcluster U5b2a2a1 in Poles) and dated as arising between 12–18 kya [...] It is also remarkable that within U5b2a1a, a Mediterranean branch precedes subcluster U5b2a1a1, which is characteristic of central and eastern European populations (Figure 1). Another U5b subcluster, U5b3, has its most likely homeland in the Italian Peninsula, from where it expanded in the Holocene along the Mediterranean coasts [11]. Hence, in general one may conclude that an initial diversification of U5b occurred in southern and central Europe, followed by the spread of a particular U5b subclusters into eastern Europe.
- 20,040.2 ± 3,208.6; CI=95% (Behar et al., 2012) haplogroup.org
- 14,938.0 ± 3,656.4; CI=95% (Behar et al., 2012) haplogroup.org
- Age: 14,676.9 ± 2,716.6; CI=95% (Behar et al., 2012) haplogroup.org
- Age: 12,651.1 ± 5,668.2; CI=95% (Behar et al., 2012) haplogroup.org
- 4,015.9 ± 3,898.7; CI=95% (Behar et al., 2012) haplogroup.org
- Matisoo-Smith EA, Gosling AL, Boocock J, Kardailsky O, Kurumilian Y, Roudesli-Chebbi S, et alMatisoo-Smith EA, Gosling AL, Boocock J, Kardailsky O, Kurumilian Y, Roudesli-Chebbi S, et al. (May 25, 2016). "A European Mitochondrial Haplotype Identified in Ancient Phoenician Remains from Carthage, North Africa". PLOS ONE. 11 (5): e0155046. Bibcode:2016PLoSO..1155046M. doi:10.1371/journal.pone.0155046. PMC 4880306. PMID 27224451.
The Young Man of Byrsa specimen dates from the late 6th century BCE, and his lineage is believed to represent early gene flow from Iberia to the Maghreb.
- Pala M, Achilli A, Olivieri A, Hooshiar Kashani B, Perego UA, Sanna D, et al. (June 2009). "Mitochondrial haplogroup U5b3: a distant echo of the epipaleolithic in Italy and the legacy of the early Sardinians" (PDF). American Journal of Human Genetics. 84 (6): 814–21. doi:10.1016/j.ajhg.2009.05.004. PMC 2694970. PMID 19500771.
- Soficaru A, Dobos A, Trinkaus E (November 2006). "Early modern humans from the Pestera Muierii, Baia de Fier, Romania". Proceedings of the National Academy of Sciences of the United States of America. 103 (46): 17196–201. Bibcode:2006PNAS..10317196S. doi:10.1073/pnas.0608443103. PMC 1859909. PMID 17085588.
- Hervella M, Svensson EM, Alberdi A, Günther T, Izagirre N, Munters AR, et al. (May 2016). "The mitogenome of a 35,000-year-old Homo sapiens from Europe supports a Palaeolithic back-migration to Africa". Scientific Reports. 6: 25501. Bibcode:2016NatSR...625501H. doi:10.1038/srep25501. PMC 4872530. PMID 27195518. "Individuals carrying haplogroup U possibly spread westward from Western Asia around 39–52 ky, reaching Europe as signaled by haplogroup U5, and North Africa signaled by haplogroup U6, which likely represents a genetic signal of a EUP return of Homo sapiens from Eurasia to North Africa. The time of the most recent common ancestor (TMRCA) for U6 was estimated to 35.3 (24.6–46.4) ky BP. [...] Our estimates of the haplogroup U6 TMRCA that incorporate ancient genomes (including PM1) set the formation of the U6 lineage back to 49.6 ky BP (95% HPD: 42–58 ky)"
- Plaza S, Calafell F, Helal A, Bouzerna N, Lefranc G, Bertranpetit J, Comas D (July 2003). "Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean" (PDF). Annals of Human Genetics. 67 (Pt 4): 312–28. doi:10.1046/j.1469-1809.2003.00039.x. PMID 12914566.
- Maca-Meyer N, González AM, Pestano J, Flores C, Larruga JM, Cabrera VM (October 2003). "Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography". BMC Genetics. 4 (1): 15. doi:10.1186/1471-2156-4-15. PMC 270091. PMID 14563219.
- Cerezo M, Černý V, Carracedo Á, Salas A (April 2011). "New insights into the Lake Chad Basin population structure revealed by high-throughput genotyping of mitochondrial DNA coding SNPs". PLOS ONE. 6 (4): e18682. Bibcode:2011PLoSO...618682C. CiteSeerX 10.1.1.291.8871. doi:10.1371/journal.pone.0018682. PMC 3080428. PMID 21533064.
- Secher B, Fregel R, Larruga JM, Cabrera VM, Endicott P, Pestano JJ, et al. (May 2014). "The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents". BMC Evolutionary Biology. 14: 109. doi:10.1186/1471-2148-14-109. PMC 4062890. PMID 24885141.
- Hernández CL, Soares P, Dugoujon JM, Novelletto A, Rodríguez JN, Rito T, et al. (2015). "Early Holocenic and Historic mtDNA African Signatures in the Iberian Peninsula: The Andalusian Region as a Paradigm". PLOS ONE. 10 (10): e0139784. Bibcode:2015PLoSO..1039784H. doi:10.1371/journal.pone.0139784. PMC 4624789. PMID 26509580.
- van de Loosdrecht M, Bouzouggar A, Humphrey L, Posth C, Barton N, Aximu-Petri A, et al. (May 2018). "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations". Science. 360 (6388): 548–552. Bibcode:2018Sci...360..548V. doi:10.1126/science.aar8380. PMID 29545507.
- Fregel R, Méndez FL, Bokbot Y, Martín-Socas D, Camalich-Massieu MD, Santana J, et al. (June 2018). "Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe". Proceedings of the National Academy of Sciences of the United States of America. 115 (26): 6774–6779. doi:10.1073/pnas.1800851115. PMC 6042094. PMID 29895688.
- Mohamed HY. Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan (PDF) (Ph.D. thesis). University of Khartoum. Retrieved 17 April 2016.
- Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, et al. (August 2004). "Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics. 5: 26. doi:10.1186/1471-2156-5-26. PMC 516768. PMID 15339343.
- Maji S, Krithika S, Vasulu TS (March 2008). "Distribution of mitochondrial DNA macrohaplogroup N in India with special reference to haplogroup R and its sub-haplogroup U." (PDF). International Journal of Human Genetics. 8 (1–2): 85–96. doi:10.1080/09723757.2008.11886022.
- Bermisheva MA, Kutuev IA, Korshunova TI, Dubova NA, Villems R, Khusnutdinova EK (2004). "[Phylogeografic analysis of mitochondrial DNA Nogays: the high level of mixture of maternal lineages from Eastern and Western Eurasia]". Molekuliarnaia Biologiia. 38 (4): 617–24. PMID 15456133.
- Derbeneva OA, Starikovskaia EB, Volod'ko NV, Wallace DC, Sukernik RI (November 2002). "[Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia]". Genetika. 38 (11): 1554–60. PMID 12500682.
- Scott R (2009). "Phylogeny of mt-hg U2a" (PDF).
- Scott R (2010). "Phylogeny of mt-hg U2b" (PDF).
- Scott R (2009). "Phylogeny of mt-hg U2c" (PDF).
- Scott R (2009). "Phylogeny of mt-hg U2d" (PDF).
- Scott R (2010). "Phylogeny of mt-hg U2e" (PDF).
- Malyarchuk B, Derenko M, Perkova M, Vanecek T (October 2008). "Mitochondrial haplogroup U2d phylogeny and distribution". Human Biology. 80 (5): 565–71. doi:10.3378/1534-6617-80.5.565. PMID 19341323.
- A. Seguin-Orlando et al. (6 November 2014). "Genomic structure in Europeans dating back at least 36,200 years". Science. 346 (6213): 1113–1118. Bibcode:2014Sci...346.1113S. doi:10.1126/science.aaa0114. PMID 25378462.CS1 maint: uses authors parameter (link)
- "DNA analysed from early European". BBC News. 2010.
- Sikora, Martin; Pitulko, Vladimir V.; Sousa, Vitor C.; Allentoft, Morten E.; Vinner, Lasse; Rasmussen, Simon; Margaryan, Ashot; De Barros Damgaard, Peter; de la Fuente Castro, Constanza; Renaud, Gabriel; Yang, Melinda; Fu, Qiaomei; Dupanloup, Isabelle; Giampoudakis, Konstantinos; Bravo Nogues, David; Rahbek, Carsten; Kroonen, Guus; Peyrot, Michäel; McColl, Hugh; Vasilyev, Sergey V.; Veselovskaya, Elizaveta; Gerasimova, Margarita; Pavlova, Elena Y.; Chasnyk, Vyacheslav G.; Nikolskiy, Pavel A.; Grebenyuk, Pavel S.; Fedorchenko, Alexander Yu.; Lebedintsev, Alexander I.; Slobodin, Sergey B.; et al. (2018). "The population history of northeastern Siberia since the Pleistocene". doi:10.1101/448829. Archived from the original on 2019-05-01. Cite journal requires
|journal=(help) - Lee EJ, Makarewicz C, Renneberg R, Harder M, Krause-Kyora B, Müller S, Ostritz S, Fehren-Schmitz L, Schreiber S, Müller J, von Wurmb-Schwark N, Nebel A (August 2012). "Emerging genetic patterns of the European Neolithic: perspectives from a late Neolithic Bell Beaker burial site in Germany". American Journal of Physical Anthropology. 148 (4): 571–9. doi:10.1002/ajpa.22074. PMID 22552938.
- Melchior L, Gilbert MT, Kivisild T, Lynnerup N, Dissing J (February 2008). "Rare mtDNA haplogroups and genetic differences in rich and poor Danish Iron-Age villages". American Journal of Physical Anthropology. 135 (2): 206–15. doi:10.1002/ajpa.20721. PMID 18046774.
- Kim K, Brenner CH, Mair VH, Lee KH, Kim JH, Gelegdorj E, et al. (July 2010). "A western Eurasian male is found in 2000-year-old elite Xiongnu cemetery in Northeast Mongolia" (PDF). American Journal of Physical Anthropology. 142 (3): 429–40. doi:10.1002/ajpa.21242. PMID 20091844.
- Scott R (2010). "Phylogeny of mt-hg U3" (PDF).
- Derenko M, Malyarchuk B, Bahmanimehr A, Denisova G, Perkova M, Farjadian S, et al. (2013). "Complete mitochondrial DNA diversity in Iranians". PLOS ONE. 8 (11): e80673. Bibcode:2013PLoSO...880673D. doi:10.1371/journal.pone.0080673. PMC 3828245. PMID 24244704.
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, et al. (November 2000). "Tracing European founder lineages in the Near Eastern mtDNA pool" (PDF). American Journal of Human Genetics. 67 (5): 1251–76. doi:10.1016/S0002-9297(07)62954-1. PMC 1288566. PMID 11032788.
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, et al. (May 2004). "Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor". American Journal of Human Genetics. 74 (5): 827–45. doi:10.1086/383236. PMC 1181978. PMID 15077202.
- Mendizabal I, Valente C, Gusmão A, Alves C, Gomes V, Goios A, et al. (January 2011). "Reconstructing the Indian origin and dispersal of the European Roma: a maternal genetic perspective". PLOS ONE. 6 (1): e15988. doi:10.1371/journal.pone.0015988. PMC 3018485. PMID 21264345.
- Kujanová M, Pereira L, Fernandes V, Pereira JB, Cerný V (October 2009). "Near eastern neolithic genetic input in a small oasis of the Egyptian Western Desert". American Journal of Physical Anthropology. 140 (2): 336–46. doi:10.1002/ajpa.21078. PMID 19425100.
- Stevanovitch A, Gilles A, Bouzaid E, Kefi R, Paris F, Gayraud RP, et al. (January 2004). "Mitochondrial DNA sequence diversity in a sedentary population from Egypt". Annals of Human Genetics. 68 (Pt 1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
- Pereira L, Cerný V, Cerezo M, Silva NM, Hájek M, Vasíková A, et al. (August 2010). "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel". European Journal of Human Genetics. 18 (8): 915–23. doi:10.1038/ejhg.2010.21. PMC 2987384. PMID 20234393.
- Salamon M, Tzur S, Arensburg B, Zias J, Nagar Y, Weiner S, Boaretto E (January 2010). "Ancient mtdna sequences and radiocarbon dating of human bones from the chalcolithic caves of wadi el-makkukh" (PDF). Mediterranean Archaeology and Archaeometry. 10 (2): 1–4. Archived from the original (PDF) on 2016-03-03.
- Budja M (December 2013). "Neolithic pots and potters in Europe: the end of 'demic diffusion' migratory model". Documenta Praehistorica. 40: 38–56. doi:10.4312/dp.40.5.
- Der Sarkissian C, Balanovsky O, Brandt G, Khartanovich V, Buzhilova A, Koshel S, et al. (2013). "Ancient DNA reveals prehistoric gene-flow from siberia in the complex human population history of North East Europe". PLOS Genetics. 9 (2): e1003296. doi:10.1371/journal.pgen.1003296. PMC 3573127. PMID 23459685.
- Haak W, Balanovsky O, Sanchez JJ, Koshel S, Zaporozhchenko V, Adler CJ, et al. (November 2010). "Ancient DNA from European early neolithic farmers reveals their near eastern affinities". PLOS Biology. 8 (11): e1000536. doi:10.1371/journal.pbio.1000536. PMC 2976717. PMID 21085689.
- Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI (April 2002). "Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis". American Journal of Human Genetics. 70 (4): 1009–14. doi:10.1086/339524. PMC 379094. PMID 11845409.
- Volodko NV, Starikovskaya EB, Mazunin IO, Eltsov NP, Naidenko PV, Wallace DC, et al. (May 2008). "Mitochondrial genome diversity in arctic Siberians, with particular reference to the evolutionary history of Beringia and Pleistocenic peopling of the Americas". American Journal of Human Genetics. 82 (5): 1084–100. doi:10.1016/j.ajhg.2008.03.019. PMC 2427195. PMID 18452887.
- "The clan of Ulrike". Maternal Ancestry. Oxford Ancestors. 2013. Archived from the original on 2017-07-15. Retrieved 2013-02-07.
- "Maternal Ancestry". Oxford Ancestors Ltd. Archived from the original on 2017-07-15. Retrieved 2013-02-07.
- Bashir M (2012). "Safeguarding Kalash heritage". Pakistan Today.
- Logan I (2014). "U4a1 Genbank Sequences".
- "The Kalash". A blog dedicated to the U4 Haplogroup.
- Scott R (2011). "Phylogeny of mt-hg U4a" (PDF).
- Scott R (2011). "Phylogeny of mt-hg U4b" (PDF).
- Scott R (2011). "Phylogeny of mt-hg U4c" (PDF).
- Scott R (2010). "Phylogeny of mt-hg U4d" (PDF).
- "Prehistoric European DNA - mtDNA & Y-DNA Haplogroup Frequencies by Period". Eupedia.
- Melchior L, Lynnerup N, Siegismund HR, Kivisild T, Dissing J (July 2010). "Genetic diversity among ancient Nordic populations". PLOS ONE. 5 (7): e11898. Bibcode:2010PLoSO...511898M. doi:10.1371/journal.pone.0011898. PMC 2912848. PMID 20689597.
- Malyarchuk B, Grzybowski T, Derenko M, Perkova M, Vanecek T, Lazur J, Gomolcak P, Tsybovsky I (August 2008). "Mitochondrial DNA phylogeny in Eastern and Western Slavs". Molecular Biology and Evolution. 25 (8): 1651–8. doi:10.1093/molbev/msn114. PMID 18477584.
- Manco J (2009). "Ancient Western Eurasian DNA of the Copper and Bronze Ages". Ancestral Journeys. Archived from the original on 2017-01-22.
- Scott R (2010). "A Comprehensive hg U Mutation List" (PDF).
- "The Oseberg women and the Gokstad man retain their genetic secrets" (PDF). Kulturhistorisk Museum. University of Oslo.
- "U7 Haplogroup Mitochondrial DNA Project". Archived from the original on 2 June 2014.
- Rudbeck L, Gilbert MT, Willerslev E, Hansen AJ, Lynnerup N, Christensen T, Dissing J (October 2005). "mtDNA analysis of human remains from an early Danish Christian cemetery". American Journal of Physical Anthropology. 128 (2): 424–9. doi:10.1002/ajpa.20294. PMID 15838837.
- Ranaweera L, Kaewsutthi S, Win Tun A, Boonyarit H, Poolsuwan S, Lertrit P (January 2014). "Mitochondrial DNA history of Sri Lankan ethnic people: their relations within the island and with the Indian subcontinental populations". Journal of Human Genetics. 59 (1): 28–36. doi:10.1038/jhg.2013.112. PMID 24196378.
- Scott R (2010). "Phylogeny of mt-hg U7" (PDF).
- Lee EJ, Steffen C, Harder M, Krause-Kyora B, Von Wurmb-Schwark N, Nebel A (January 2012). "An ancient DNA perspective on the Iron Age "princely burials" from Baden-Wiirttemberg, Germany". American Journal of Physical Anthropology. 147: 190. doi:10.1002/ajpa.22033.
- Berglund N (26 March 2007). "Viking woman had roots near the Black Sea -". Aftenposten - News in English. Aftenposten.no. Archived from the original on 2010-08-26. Retrieved 11 April 2010.
- Farrell JJ, Al-Ali AK, Farrer LA, Al-Nafaie AN, Al-Rubaish AM, Melista E, et al. "The Saudi Arabian Genome Reveals a Two Step Out-of-Africa Migration". American Society of Human Genetics (ASHG).
- Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, Pennarun E, et al. (November 2004). "Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears". American Journal of Human Genetics. 75 (5): 752–70. doi:10.1086/425161. PMC 1182106. PMID 15457403.
Further reading
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli EL, Silva NM, Kivisild T, Torroni A, Villems R (April 2012). "A "Copernican" reassessment of the human mitochondrial DNA tree from its root". American Journal of Human Genetics. 90 (4): 675–84. doi:10.1016/j.ajhg.2012.03.002. PMC 3322232. PMID 22482806.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- Mannis van Oven's Phylotree
- Haplogroup U
- Danish Demes Regional DNA Project: mtDNA Haplogroup U
- Spread of Haplogroup U, from National Geographic