Genetic studies on Serbs
Genetic studies on Serbs show close affinity to other neighboring South Slavs.[1]
Y-DNA
Y-chromosomal haplogroups identified among the Serbs from Serbia and near countries are the following with respective percentages: I2a (36.6[2]-42%[3]), E1b1b (16.5[3]-18.2%[2]), R1a (14.9[2]-15%[3]), R1b (5[2]-6%[3]), I1 (1.5[3]-7.6%[2]), J2b (4.5[3]-4.9%[2]), J2a (4[4]-4.5%[3]), J1 (1[4]-4.5%[3]), G2a (1.5[3]-5.8%[4]), and several other uncommon haplogroups with lesser frequencies.[3][5][6][7]
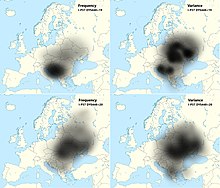
I2a-P37.2 is the most prevailing haplogroup, accounting for one-third of Serbians. It is represented by four sub-clusters I-PH908 (25.08%), I2a1b3-L621 (7.59%), I2-CTS10228 (3.63%) and I2-M223 (0.33%).[2] Older research considered that the high frequency of this subclade in the South Slavic-speaking populations to be the result of a "pre-Slavic" paleolithic settlement in the region, the research by O.M. Utevska (2017) confirmed that the haplogroup STR haplotypes have the highest diversity in Ukraine, with ancestral STR marker result "DYS448=20" comprising "Dnieper-Carpathian" cluster, while younger derived result "DYS448=19" comprising the "Balkan cluster" which is predominant among the South Slavs.[8] This "Balkan cluster" also has the highest variance in Ukraine, which indicates that the very high frequency in the Western Balkan is because of a founder effect.[8] Utevska calculated that the STR cluster divergence and its secondary expansion from the middle reaches of the Dnieper river or from Eastern Carpathians towards the Balkan peninsula happened approximately 2,860 ± 730 years ago, relating it to the times before Slavs, but much after the decline of the Cucuteni–Trypillia culture.[8] More specifically, the "Balkan cluster" is represented by a single SNP, I-PH908, known as I2a1a2b1a1a1c in ISOGG phylogenetic tree (2019), and according to YFull YTree it formed and had TMRCA approximately 1,850-1,700 YBP (2nd-3rd century AD).[9] Although I-L621 it is dominant among the modern Slavic peoples on the territory of the former Balkan provinces of the Roman Empire, until now it was not found among the samples from the Roman period and is almost absent in contemporary population of Italy.[10] It was found in the skeletal remains with artifacts, indicating leaders, of Hungarian conquerors of the Carpathian Basin from the 9th century, part of Western Eurasian-Slavic component of the Hungarians.[10] According to Fóthi et al. (2020), the distribution of ancestral subclades like of I-CTS10228 among contemporary carriers indicates a rapid expansion from Southeastern Poland, is mainly related to the Slavs, and the "largest demographic explosion occurred in the Balkans".[10]
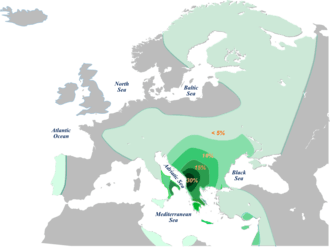
E1b1b-M215 is the second most prevailing haplogroup amongst Serbs, accounting for nearly one-fifth of Serbians. It is represented by four sub-clusters E-V13 (17.49%), E1b1b-V22 (0.33%), and E1b1b-M123 (0.33%).[2] In Southeast Europe, its frequency peaks at the southeastern edge of the region and its variance peaks in the region's southwest. Although its frequency is very high in Kosovar Albanians (46%) and Macedonian Romani (30%), this phenomenon is of a focal rather than a clinal nature, most likely being a consequence of genetic drift.[5] E-V13 is also high amongst Albanians in North Macedonia (34%) and Albanians in Albania (24%), as well as ethnic Macedonians, Romanians, and Greeks. It is found at low to moderate frequencies in most Slavic populations. However, amongst South Slavs, it is quite common. It is found in 27% of Montenegrins, 22% of Macedonians, and 18% of Bulgarians, all Slavic peoples. Moderate frequencies of E-V13 are also found in Italy and western Anatolia.[5][7] In most of Central Europe (Hungary, Austria, Switzerland, Ukraine, Slovakia), it is found at low to moderate frequencies of 7-10%, in both R1a (Slavic) and R1b (Germanic/Celtic) dominated populations. It likely originated in the Balkans, Greece, or the Carpathian Basin 9000 YBP or shortly before its arrival in Europe during the Neolithic. Its ancestral haplogroup, E1b1b1a-M78, is of northeast African origin.[7]
R1a1-M17 accounts for about one-seventh to one-sixth of Serbian Y-chromosomes. It is represented by four sub-clusters R1a (10.89%), R1a-M458 (2.31%), R1a-YP4278 (1.32%), and R1a-Y2613 (0.33%).[2] Its frequency peaks in Ukraine (54.0%).[5] It is the most predominant haplogroup in the general Slavic paternal gene pool. The variance of R1a1 in the Balkans might have been enhanced by infiltrations of Indo-European speaking peoples between 2000 and 1000 BC, and by the Slavic migrations to the region in the early Middle Ages.[5][6] A descendant lineage of R1a1-M17, R1a1a7-M458, has the highest frequency in Central and Southern Poland.[11]
R1b1b2-M269 is moderately represented among Serbian males (6–10%), 10% in Serbia (Balaresque et al. 2010),[12] with subclade M269* (xL23) 4.4% in Serbia, 5.1% in Macedonia, 7.9% in Kosovo. The highest frequency in the central Balkans (Myres et al. 2010).[13] It has its frequency peak in Western Europe (90% in Wales), but a high frequency is also found in Central Europe among the West Slavs (Poles, Czechs, Slovaks) and Hungarians as well as in the Caucasus among the Ossetians (43%).[5] It was introduced to Europe by farmers migrating from western Anatolia, probably about 7500 YBP. Serb bearers of this haplogroup are in the same cluster as Central and East European ones, as indicated by the frequency distributions of its sub-haplogroups with respect to total R-M269. The other two clusters comprise, respectively, West Europeans and a group of populations from Greece, Turkey, the Caucasus and the Circum-Uralic region.[14]
J2b-M102 and J2a1b1-M92 have low frequencies among the Serbs (6–9% combined). Various other lineages of haplogroup J2-M172 are found throughout the Balkans, all with low frequencies. Haplogroup J and all its descendants originated in the Middle East. It is proposed that the Balkan Mesolithic foragers, bearers of I-P37.2 and E-V13, adopted farming from the initial J2 agriculturalists who colonized the region about 7000 to 8000 YBP, transmitting the Neolithic cultural package.[7]
I1-M253 is also found in low frequencies (1.5-7.6%) and is represented by three sub-clusters I1-P109 (5.28%), I1 (1.32%), and I1-Z63 (0.99%).[2]
An analysis of molecular variance based on Y-chromosomal STRs showed that Slavs can be divided into two groups: one encompassing West Slavs, East Slavs, Slovenes, and western Croats, and the other – all remaining Southern Slavs. Croats from northern Croatia (Zagreb region) fell into the second group. This distinction could be explained by a genetic contribution of pre-Slavic Balkan populations to the genetic heritage of some South Slavs belonging to the group.[15] Principal component analysis of Y-chromosomal haplogroup frequencies among the three ethnic groups in Bosnia and Herzegovina, Serbs, Croats, and Bosniaks, showed that Serbs and Bosniaks are genetically closer to each other than either of them is to Croats.[6]
Tables
2000s
| Population | Samples | Source | E3b* | E3b1 | E3b1-α | E3b2 | E3b3 | G | J | J2e* | J2e1 | J2* | J2f* | J2f1 | F* | H1 | I* | I1a | xM26 | I1c | K*(xP) | R1b | R1a | Q* | P*(xQ,R1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbians, Belgrade | 113 | Peričić et al. (2005)[16] | 0 | 1.77 | 18.58 | 0 | 0.90 | 0 | 0 | 4.40 | 0.90 | 0 | 0 | 2.70 | 0 | 0.90 | 1.77 | 5.31 | 29.20 | 0 | 7.08 | 10.62 | 15.93 | 0 | 0 |
| Population | Samples | Source | E3b* | E3b1 | G | I1a | I1b* | I* | I1c | J1 | J2* | J2e | J2f* | J2f1 | F* | K* | R1a1 | R1b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbs, B&H | 81 | Marjanović et al. (2005)[17] | 2.5 | 19.8 | 1.2 | 2.5 | 30.9 | 1.2 | 1.2 | 0 | 2.5 | 6.2 | 0 | 0 | 4.9 | 7.4 | 13.6 | 6.2 |
| Population | Samples | Source | E*1b1b1 | E1b1b1a2 | G2a* | I1* | I2* | I2a1* | I2b1 | J1* | J2a1k | J2b* | J2b2 | N1 | R1a1* | R1b1b2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbs, B&H | 81 | Battaglia et al. (2008)[18] | 2.5 | 19.8 | 1.2 | 2.5 | 2.5 | 34.6 | 1.2 | 1.2 | 2.5 | 3.7 | 2.5 | 6.2 | 13.6 | 6.2 |
2010s
| Population | Samples | Source | I2a | R1a | E1b1b | I1 | R1b | J2b | J2a | J1 | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbians | 179 | Mirabal et al. (2010)[19][20] | 39.8 | 14.6 | 17.3 | 5.8 | 4.8 | 1.6 | 2.6 | 0.5 | 13 |
| Population | Samples | Source | I2a | R1a | E1b1b | I1 | G | R1b | J2b | J2a | J1 | N | Q | I2+ | other | unknown |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbians | 103 | Reguiero et al. (2012)[21] | 29,1 | 20,4 | 18,5 | 7,8 | 5,8 | 7,8 | 2,9 | 4 | 1 | 1,9 | 0 | 1 | 0 | 0 |
| Serbs, Aleksandrovac | 85 | Todorović et al. (2014)[22] | 35,29 | 21,17 | 15,29 | 4,70 | 10,58 | 1,17 | 4,70 | 2,35 | 2,35 | 0 | 1,17 | 0 | 0 | 1,17 |
| Population | Samples | Source | I2a | R1a | E1b1b | E1b1 | I1 | R1b | J2b | J2a | J1 | T | G2a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbians | 257 | Scorrano et al. (2017)[3] | 42 | 15 | 12 | 4.5 | 1.5 | 6 | 4.5 | 4.5 | 4.5 | 4.5 | 1.5 |
| Population | Samples | Source | I2a | E1b1b | R1a | I1 | R1b | J2b | J2a | J1 | N | G2a | Q | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serbia | 203 | Zgonjanin et al. (2017)[23] | 32.1 | 22.5 | 16.3 | 7.7 | 4.8 | 2.9 | 2.4 | 2.9 | 3.8 | 1.9 | 1.9 | 0 |
| North Serbia | 68 | Zgonjanin et al. (2017)[23] | 30.4 | 21.7 | 18.8 | 4.3 | 5.8 | 4.3 | 1.4 | 1.4 | 7.2 | 4.3 | 0 | 0 |
| Central Serbia | 68 | Zgonjanin et al. (2017)[23] | 33.8 | 16.9 | 16.9 | 11.3 | 5.6 | 2.8 | 4.2 | 2.8 | 1.4 | 0 | 2.8 | 1.4 |
| South Serbia | 67 | Zgonjanin et al. (2017)[23] | 31.8 | 29.0 | 13.0 | 7.2 | 2.9 | 1.4 | 1.4 | 4.5 | 2.9 | 1.4 | 2.9 | 1.4 |
| Population | Samples | Source | I2a | E1b1b | R1a | I1 | R1b | J2b | J (J1 & J2a) | Other |
|---|---|---|---|---|---|---|---|---|---|---|
| Serbs (Serbia, Montenegro, BiH, Croatia) | 303 | Kačar et al. (2019)[2] | 36.6 | 18.2 | 14.9 | 7.6 | 5 | 4.9 | 5.9 | 6.9 |
mtDNA
According to Davidovic et al. (2014) study of Mitochondrial DNA in 139 samples in Serbia are present "mtDNA lineages predominantly found within the Slavic gene pool (U4a2a*, U4a2a1, U4a2c, U4a2g, HV10), supporting a common Slavic origin, but also lineages that may have originated within the southern Europe (H5*, H5e1, H5a1v) and the Balkan Peninsula in particular (H6a2b and L2a1k)".[24] According to 2017 study on haplogroup U diversity "putative Balkan-specific lineages (e.g. U1a1c2, U4c1b1, U5b3j, K1a4l and K1a13a1) and lineages shared among Serbians (South Slavs) and West and East Slavs were detected (e.g. U2e1b1, U2e2a1d, U4a2a, U4a2c, U4a2g1, U4d2b and U5b1a1). The exceptional diversity of maternal lineages found in Serbians may be associated with the genetic impact of both autochthonous pre-Slavic Balkan populations whose mtDNA gene pool was affected by migrations of various populations over time (e.g. Bronze Age pastoralists) and Slavic and Germanic newcomers in the early Middle Ages".[25] The 2020 study of 226 samples mitochondrial genome data of Serbian population "supported more pronounced genetic differentiation among Serbians and two Slavic populations (Russians and Poles) as well as expansion of the Serbian population after the Last Glacial Maximum and during the Migration period (fourth to ninth century A.D.)".[26]
Autosomal DNA
According to 2013 autosomal IBD survey "of recent genealogical ancestry over the past 3,000 years at a continental scale", the speakers of Serbo-Croatian language share a very high number of common ancestors dated to the migration period approximately 1,500 years ago with Poland and Romania-Bulgaria cluster among others in Eastern Europe. It is concluded to be caused by the Hunnic and Slavic expansion, which was a "relatively small population that expanded over a large geographic area", particularly "the expansion of the Slavic populations into regions of low population density beginning in the sixth century" and that it is "highly coincident with the modern distribution of Slavic languages".[27] The 2015 IBD analysis found that the South Slavs have lower proximity to Greeks than with East Slavs and West Slavs, and "even patterns of IBD sharing among East-West Slavs–'inter-Slavic' populations (Hungarians, Romanians and Gagauz)–and South Slavs, i.e. across an area of assumed historic movements of people including Slavs". The slight peak of shared IBD segments between South and East-West Slavs suggests a shared "Slavonic-time ancestry".[28] The 2014 IBD analysis comparison of Western Balkan and Middle Eastern populations found negligible gene flow between 16th and 19th century during the Islamization of the Balkans.[29]
According to a 2014 autosomal analysis of Western Balkan, the Serbian population shows genetic uniformity with other South Slavic populations, but the "Serbians and Montenegrins have an intermediate position on PCA plot and on Fst–based network among other Western Balkan populations".[29] In the 2015 analysis, Serbians were again in the middle of a Western South Slavic cluster (Croatians, Bosnians and Slovenians) and Eastern South Slavic cluster (Macedonians and Bulgarians). The western cluster has an inclination toward Hungarians, Czechs, and Slovaks, while the eastern cluster toward Romanians and some extent Greeks.[28] The studies also found very high correlation between genetic, geographic and linguistic distances of Balto-Slavic populations.[29][28]
A genetic study of Sorbs showed that they share greatest affinity with Poles, while results were compared to Serbia and Montenegro due to historical hypotheses of common origin, showing that "The Sorbs were most different to Serbia and Montenegro, likely reflecting the considerable geographical distance between the two populations."[30]
Physical anthropology
According to Serbian physical anthropologist Živko Mikić, the medieval population of Serbia developed a phenotype that represented a mixture of Slavic and indigenous Balkan Dinaric traits. Mikić argues that the Dinaric traits, such as brachycephaly and a bigger average height, have been since then becoming predominant over the Slavic traits among Serbs.[31]
Gallery
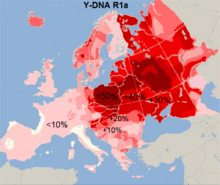 R1a distribution
R1a distribution- R1b distribution
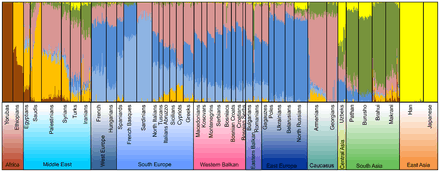 Admixture analysis of autosomal SNPs in a global context on the resolution level of 7 assumed ancestral populations per Kovačević et al. (2014)
Admixture analysis of autosomal SNPs in a global context on the resolution level of 7 assumed ancestral populations per Kovačević et al. (2014) Principal component (PC) analysis of the variation of autosomal SNPs in Western Balkan populations in Eurasian context per Kovačević et al. (2014)
Principal component (PC) analysis of the variation of autosomal SNPs in Western Balkan populations in Eurasian context per Kovačević et al. (2014)- Admixture analysis on the resolution level of 6 assumed ancestral populations per Kushniarevich et al. (2015)
 PC1vsPC2 plot based on whole genome SNP data per Kushniarevich et al. (2015)
PC1vsPC2 plot based on whole genome SNP data per Kushniarevich et al. (2015)
See also
| Part of a series of articles on |
| Serbs |
|---|
 |
|
Native communities
|
|
|
|
|
|
Related groups |
References
- Novembre, J; Johnson, T; Bryc, K; et al. (2008). "(November 2008), "Genes mirror geography within Europe". Nature. 456 (7218): 98–101. doi:10.1038/nature07331. PMC 2735096. PMID 18758442.
- Kačar et al. 2019.
- Scorrano et al. 2017.
- Regueiro et al. 2012.
- Peričić et al. 2005
- Marjanović et al. 2005
- Battaglia et al. 2008
- O.M. Utevska (2017). Генофонд українців за різними системами генетичних маркерів: походження і місце на європейському генетичному просторі [The gene pool of Ukrainians revealed by different systems of genetic markers: the origin and statement in Europe] (PhD) (in Ukrainian). National Research Center for Radiation Medicine of National Academy of Sciences of Ukraine. pp. 219–226, 302.
- "I-PH908 YTree v8.06.01". YFull.com. 27 June 2020. Retrieved 17 July 2020.
- Fóthi, E.; Gonzalez, A.; Fehér, T.; et al. (2020), "Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes", Archaeological and Anthropological Sciences, 12 (1), doi:10.1007/s12520-019-00996-0
- Underhill PA, Myres NM, Rootsi S, et al. (April 2010). "Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a". European Journal of Human Genetics. 18 (4): 479–84. doi:10.1038/ejhg.2009.194. PMC 2987245. PMID 19888303.
- Balaresque et al. 2010
- Myres et al. 2010
- Myres NM, Rootsi S, Lin AA, et al. (January 2011). "A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe". European Journal of Human Genetics. 19 (1): 95–101. doi:10.1038/ejhg.2010.146. PMC 3039512. PMID 20736979.
- Rebała, K; Mikulich, AI; Tsybovsky, IS; Siváková, D; Dzupinková, Z; Szczerkowska-Dobosz, A; Szczerkowska, Z (2007). "Y-STR variation among Slavs: Evidence for the Slavic homeland in the middle Dnieper basin". Journal of Human Genetics. 52 (5): 406–14. doi:10.1007/s10038-007-0125-6. PMID 17364156.
- Peričić et al. 2005.
- Marjanović et al. 2005.
- Battaglia et al. 2008.
- Mirabal et al. 2010.
- Šehović et al. 2018.
- Todorović et al. 2014a, p. 259, citing Reguiero et al. 2012
- Todorović et al. 2014a, p. 251.
- Zgonjanin et al. 2017.
- Davidovic et al. 2015.
- Davidovic et al. 2017.
- Davidovic et al. 2020.
- P. Ralph (2013). "The Geography of Recent Genetic Ancestry across Europe". PLOS Biology. 11 (5): e105090. doi:10.1371/journal.pbio.1001555. PMC 3646727. PMID 23667324.
- A. Kushniarevich (2015). "Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data". PLOS One. 10 (9): e0135820. doi:10.1371/journal.pone.0135820. PMC 4558026. PMID 26332464.
- L. Kovačević (2014). "Standing at the Gateway to Europe - The Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers". PLOS One. 9 (8): e105090. doi:10.1371/journal.pone.0105090. PMC 4141785. PMID 25148043.
- Veeramah, KR; Tönjes, A; Kovacs, P; Gross, A; Wegmann, D; Geary, P; Gasperikova, D; Klimes, I; Scholz, M; Novembre, J; Stumvoll, M. "Genetic variation in the Sorbs of eastern Germany in the context of broader European genetic diversity". Eur J Hum Genet. 19: 995–1001. doi:10.1038/ejhg.2011.65. PMC 3179365. PMID 21559053.
- Mikić Ž (1994). "Beitrag zur Anthropologie der Slawen auf dem mittleren und westlichen Balkan". Balcanica". (Belgrade: The Institute for Balkan Studies of the Serbian Academy of Sciences and Arts). 25: 99–109.
Sources
- Battaglia, Vincenza; et al. (2008). "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe". European Journal of Human Genetics. 17 (6): 820–30. doi:10.1038/ejhg.2008.249. PMC 2947100. PMID 19107149.
- Bosch, E.; Calafell, F.; González-Neira, A.; Flaiz, C; Mateu, E; Scheil, HG; Huckenbeck, W; Efremovska, L; et al. (2006). "Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns" (PDF). Annals of Human Genetics. 70 (Pt 4): 459–87. doi:10.1111/j.1469-1809.2005.00251.x. PMID 16759179.
- Cvjetan, S; Tolk, HV; Lauc, LB; Colak, I; Dordević, D; Efremovska, L; Janićijević, B; Kvesić, A; et al. (2004). "Frequencies of mtDNA haplogroups in southeastern Europe--Croatians, Bosnians and Herzegovinians, Serbians, Macedonians and Macedonian Romani". Collegium Antropologicum. 28 (1): 193–8. PMID 15636075.
- Davidović, S. (2015). "Mitochondrial DNA perspective of Serbian genetic diversity". Am J Phys Anthropol. 156 (3): 449–65. doi:10.1002/ajpa.22670. PMID 25418795.
- Davidović, S. (2017). "Mitochondrial super-haplogroup U diversity in Serbians". Annals of Human biology. 44 (5): 408–418. doi:10.1080/03014460.2017.1287954. PMID 28140657.
- Davidović, S.; Malyarchuk, B.; Grzybowski, T. (2020). "Complete mitogenome data for the Serbian population: the contribution to high-quality forensic databases". Int J Legal Med. doi:10.1007/s00414-020-02324-x.
- Kačar, Tamara (2019). "Y chromosome genetic data defined by 23 short tandem repeats in a Serbian population on the Balkan Peninsula". Annals of Human Biology. 46 (1): 1–21. doi:10.1080/03014460.2019.1584242.
- Kovačević, Lejla (2014). "Standing at the Gateway to Europe - The Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers". PLOS One. 9 (8): e105090. doi:10.1371/journal.pone.0105090. PMC 4141785. PMID 25148043.
- Marjanović, D; Fornarino, S; Montagna, S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Annals of Human Genetics. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Mirabal, S.; Varljen, T.; Gayden, T. (July 2010). "Human Y-chromosome short tandem repeats: A tale of acculturation and migrations as mechanisms for the diffusion of agriculture in the Balkan Peninsula". American Journal of Physical Anthropology. 142 (3): 380–390. doi:10.1002/ajpa.21235. PMID 20091845.
- Peričić, M; Lauc, LB; Klarić, IM; et al. (October 2005). "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations". Mol. Biol. Evol. 22 (10): 1964–75. doi:10.1093/molbev/msi185. PMID 15944443.
- Regueiro, M. (2012). "High levels of Paleolithic Y-chromosome lineages characterize Serbia". Gene. 498 (1): 59–67. doi:10.1016/j.gene.2012.01.030. PMID 22310393.
- Scorrano, Gabriele (2017). "The Genetic Landscape of Serbian Populations through Mitochondrial DNA Sequencing and Non-Recombining Region of the Y Chromosome Microsatellites". Collegium antropologicum. 41 (3): 275–296.
- Šehović, Emir (2018). "A glance of genetic relations in the Balkan populations utilizing network analysis based on in silico assigned Y-DNA haplogroups". Anthropological Review. 81 (3): 252–268. doi:10.2478/anre-2018-0021.
- Todorović, I.; Vučetić-Dragović, A.; Marić, A. (2014). "Непосредни резултати нових мултидисциплинарних етногенетских истраживања Срба и становништва Србије (на примеру Александровачке жупе)" (PDF). Glasnik Etnografskog Instituta SANU. 62 (1): 245–258. doi:10.2298/GEI1401245T. Archived from the original (PDF) on 2017-09-20.
- Todorović, I. (2013). "Нове могућности етногенетских проучавања становништва Србије" (PDF). Glasnik Etnografskog Instituta SANU. 61 (1): 149–159. doi:10.2298/GEI1301149T. Archived from the original (PDF) on 2015-12-27.
- Todorović, I.; Vučetić-Dragović, A.; Marić, A. (2014). "Компаративни аналитички осврт на најновија генетска истраживања порекла Срба и становништва Србије – етнолошка перспектива" (PDF). Glasnik Etnografskog Instituta SANU. 62 (2): 99–111. doi:10.2298/GEI1402099T.
- Veselinović, Igor S. (March 2008). "Allele frequencies and population data for 17 Y-chromosome STR loci in a Serbian population sample from Vojvodina province". Forensic Science International. 176 (2–3): 23–28. doi:10.1016/j.forsciint.2007.04.003. PMID 17482396.
- Zgonjanin, Dragana (2017). "Genetic characterization of 27 Y-STR loci with the Yfiler® Plus kit in the population of Serbia". Forensic Science International: Genetics. 31. doi:10.1016/j.fsigen.2017.07.013.
Further reading
- Kushniarevich, Alena (September 2, 2015). "Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data". PLOS One. 10 (9): e0135820. doi:10.1371/journal.pone.0135820. PMC 4558026. PMID 26332464.
- Kovacevic, Lejla; Tambets, Kristiina; Ilumäe, Anne-Mai; Kushniarevich, Alena; Yunusbayev, Bayazit; Solnik, Anu; Bego, Tamer; Primorac, Dragan; Skaro, Vedrana (2014-08-22). "Standing at the Gateway to Europe - The Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers". PLOS One. 9 (8): e105090. doi:10.1371/journal.pone.0105090. ISSN 1932-6203. PMC 4141785. PMID 25148043.
- "The Genomic History Of Southeastern Europe". bioRxiv 10.1101/135616.
- Myres, Natalie; Rootsi, Siiri; Lin, Alice A; Järve, Mari; King, Roy J; Kutuev, Ildus; Cabrera, Vicente M; Khusnutdinova, Elza K; et al. (2010). "A major Y-chromosome haplogroup R1b Holocene effect in Central and Western Europe". European Journal of Human Genetics. 19 (1): 95–101. doi:10.1038/ejhg.2010.146. PMC 3039512. PMID 20736979.
- Balaresque, Patricia; Bowden, Georgina R.; Adams, Susan M.; Leung, Ho-Yee; King, Turi E.; et al. (2010). Penny, David (ed.). "A Predominantly Neolithic Origin for European Paternal Lineages". PLOS Biology. 8 (1): e1000285. doi:10.1371/journal.pbio.1000285. PMC 2799514. PMID 20087410.
External links
- Serbian DNA Project
- Poreklo - Society of genetic genealogy