Genetic studies on Bosniaks
As with all modern European nations, a large degree of 'biological continuity' exists between the Bosniaks and their ancient predecessors with Bosniak Y chromosomal lineages testifying to predominantly Paleolithic European ancestry.[1][2] A majority (>67%) of Bosniaks belong to one of the three major European Y-DNA haplogroups: I2 (43.50%), R1a (15.3%) and R1b (3.5%), while a minority belongs to less frequently occurring haplogroups E-V13 (12.90%) and J2 (8.7%), along with other more rare lineages.[3]
Studies based on bi-allelic markers of the NRY (non-recombining region of the Y-chromosome) have shown the three main ethnic groups of Bosnia and Herzegovina (Bosniaks, Bosnian Serbs and Bosnian Croats) to share, in spite of some quantitative differences, a large fraction of the same ancient gene pool distinct for the region.[4] Analysis of autosomal STRs have moreover revealed no significant difference between the population of Bosnia and Herzegovina and neighbouring populations.[5]
Autosomal DNA of Bosniaks in Bosnia and Herzegovina
According to 2013 autosomal IBD survey "of recent genealogical ancestry over the past 3,000 years at a continental scale", the speakers of Serbo-Croatian language share a very high number of common ancestors dated to the migration period approximately 1,500 years ago with Poland and Romania-Bulgaria cluster among others in Eastern Europe. It is concluded to be caused by the Hunnic and Slavic expansion, which was a "relatively small population that expanded over a large geographic area", particularly "the expansion of the Slavic populations into regions of low population density beginning in the sixth century" and that it is "highly coincident with the modern distribution of Slavic languages".[6] The 2015 IBD analysis found that the South Slavs have lower proximity to Greeks than with East Slavs and West Slavs, and "even patterns of IBD sharing among East-West Slavs–'inter-Slavic' populations (Hungarians, Romanians and Gagauz)–and South Slavs, i.e. across an area of assumed historic movements of people including Slavs". The slight peak of shared IBD segments between South and East-West Slavs suggests a shared "Slavonic-time ancestry".[7]
An 2014 autosomal analysis study of 90 samples showed that Western Balkan populations had a genetic uniformity, intermediate between South Europe and Eastern Europe, in line with their geographic location. According to the same study, Bosnians (together with Croatians) are by autosomal DNA closest to East European populations and overlap mostly with Hungarians.[8] In the 2015 analysis, Bosnians formed a western South Slavic cluster with the Croatians and Slovenians in comparison to eastern cluster formed by Macedonians and Bulgarians with Serbians in the middle. The western cluster (Bosnians included) has an inclination toward Hungarians, Czechs, and Slovaks, while the eastern cluster toward Romanians and some extent Greeks.[7] Based on analysis of IBD sharing, Middle Eastern populations most likely did not contribute to genetics in Islamicized populations in the Western Balkans, including Bosniaks, as these share similar patterns with neighboring Christian populations.[8]
Y-DNA frequency of Bosniaks in Bosnia and Herzegovina
Y-DNA studies on Bosniaks (in Bosnia and Herzegovina) show close affinity to other neighboring South Slavs.[9] Y-DNA results show notable frequencies of I2 with 43.50% (especially its subclade I2-CTS10228>S17250+), R1a with 15.30% (mostly its two subclades R1a-CTS1211+ and R1a-M458+), E-V13 with 12.90% and J-M410 with 8.7%.
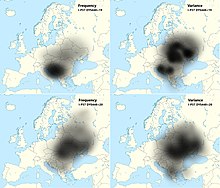
The frequency of haplogroup I2, especially its subclade I2-CTS10228 and its variance, peaks over a large geographic area covering Bosnia and Herzegovina, Croatia, Serbia, Montenegro, North Macedonia, Albania, Bulgaria, Romania, Moldova, Slovakia, Ukraine, Belarus, Poland and western Russia. In comparison to older research which argued a prehistoric autochthonous origin of the haplogroup I2 in western Balkans,[10][11][nb 1] the most recent research by O. M. Utevska (2017) found the haplogroups STR haplotypes have the highest diversity in Ukraine, with ancestral STR marker result DYS448=20 comprising "Dnieper-Carpathian" cluster (modal Y-STR for I2-CTS10228>S17250), while younger derived result DYS448=19 (modal Y-STR for I2-CTS10228>S17250>PH908) comprising the "Balkan cluster" which is predominant among the South Slavs[17], but can also be found in East and West Slavic populations.[18] The clusters divergence and gradual expansion from the Carpathians in the direction of the Balkan peninsula happened approximately 2,860 ± 730 years ago, coinciding with the Slavic migration. The lack of diversity of DYS448=19 haplotypes in the Western Balkan also indicate a founder effect.[17] Although it is considered that I-L621 might have been present in the Cucuteni–Trypillia culture,[19] until now was only found G2a,[20] and another subclade I2a1a1-CTS595 was present in the Baden culture of the Calcholitic Carpathian Basin.[19][21] Although it is dominant among the modern Slavic peoples on the territory of the former Balkan provinces of the Roman Empire, until now it was not found among the samples from the Roman period and is almost absent in contemporary population of Italy.[22] It was found in the skeletal remains with artifacts, indicating leaders, of Hungarian conquerors of the Carpathian Basin from the 9th century, part of Western Eurasian-Slavic component of the Hungarians.[22][19] According to Fóthi et al. (2020), the distribution of ancestral subclades like of I-CTS10228 among contemporary carriers indicates a rapid expansion from Southeastern Poland, is mainly related to the Slavs, and the "largest demographic explosion occurred in the Balkans".[22]
Y-DNA studies done for the majority Bosniak populated city of Zenica and Tuzla Canton, shows however a drastic increase of the two major haplogroups I2 and R1a. Haplogroup I2 scores 52.20% in Zenica (Peričić et al., 2005) and 47% in Tuzla Canton (Dogan et al., 2016), while R1a increases up to 24.60% and 23% in respective region.[23][24] Principal component analysis of Y-chromosomal haplogroup frequencies among the three ethnic groups in Bosnia and Herzegovina, Serbs, Croats, and Bosniaks, showed that Bosnian Serbs and Bosniaks are by Y-DNA closer to each other than either of them is to Bosnian Croats.[25]
Frequencies by region
| Region | Samples | Source[26] | E1b1b | G | I1 | I2a1 | J1 | J2 | F* | K* | R1a | R1b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bosnia and Herzegovina | 85 | Marjanović et al. (2005) | 12.94%=(11/85) | 3.53%=(3/85) | 4.71%(4/85) | 43.53%=(37/85) | 2.35%=(2/85) | 8.7% | 3.53%(3/85) | 1.18%=(1/85) | 15.29%=(13/85) | 3.53%(3/85) |
| Region | Samples | Source[27] | E1b1b | G | I1 | I2a1 | J2a | J2b | N | R1a | R1b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bosnia and Herzegovina | 100 | Doğan et al. (2017) | 17% | 1% | 4% | 49% | 5% | 2% | 1% | 17% | 4% |
| Region | Samples | Source[28] | E1b1b | G | I1 | I2a1 | R1a | R1b |
|---|---|---|---|---|---|---|---|---|
| Zenica | 69 | Peričić et al. (2005) | 10.15%=(7/69) | 4.35%=(3/69) | 1.45%=(1/69) | 52.17%=(36/69) | 24.64%=(17/69) | 1.45%=(1/69) |
| Region | Samples | Source[29] | E1b1b | G | I1 | I2a1 | J2 | N2 | Q | R1a | R1b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tuzla Canton | 100 | Dogan et al. (2016) | 7% | 2% | 4% | 47% | 7% | 4% | 1% | 23% | 5% |
Mitochondrial DNA
Genetically, on the maternal X chromosome line, a majority (>75%) of Bosnians belong to three of the eleven major European mtDNA haplogroups - H (47.92%), U (19.44%) and J (6.94%), while a large minority (>25) belongs to other rare mitochondrial lineages.[30] The mtDNA studies shows that the Bosnian population partly share similarities with other Southern European populations (especially with mtDNA haplogroups such as pre-HV (today known as mtDNA haplogroup R0), HV2 and U1), but are for the mostly featured by a huge combination of mtDNA subclusters that indicates a consanguinity with Central and Eastern Europeans, such as modern German, West Slavic, East Slavic and Finno-Ugric populations. There is especially the observed similarity between Bosnian, Russian and Finnish samples (with mtDNA subclusters such as U5b1, Z, H-16354, H-16263, U5b-16192-16311 and U5a-16114A). The huge differentiation between Bosnian and Slovene samples of mtDNA subclusters that are also observed in Central and Eastern Europe, may suggests a broader genetic heterogeneity among the Slavs that settled the Western Balkans during the early Middle ages.[30] The 2019 study of ethnic groups of Tuzla Canton of Bosnia and Herzegovina (Bosniaks, Croats and Serbs) found "close gene similarity among maternal gene pools of the ethnic groups of Tuzla Canton", which is "suggesting similar effects of the paternal and maternal gene flows on genetic structure of the three main ethnic groups of modern Bosnia and Herzegovina".[31]
References
- Marjanović, D; Fornarino, S; Montagna, S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Annals of Human Genetics. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Peričić, M; Barać Lauc, L; Martinović, I; et al. (2005). "High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations" (PDF). Molecular Biology and Evolution. 22 (10): 1964–75. doi:10.1093/molbev/msi185. PMID 15944443.
- Marjanović, D; Fornarino, S; Montagna, S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Annals of Human Genetics. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Damir, Marjanović / International Congress Series 1288 (2006) 243-245; et al. (2006). "Preliminary population study at fifteen autosomal and twelve Y-chromosome short tandem repeat loci in the representative sample of multinational Bosnia and Herzegovina residents" (PDF). Institute for Genetic Engineering and Biotechnology: 244.
- Damir, Marjanović / International Congress Series 1288 (2006) 243-245; et al. (2006). "Preliminary population study at fifteen autosomal and twelve Y-chromosome short tandem repeat loci in the representative sample of multinational Bosnia and Herzegovina residents" (PDF). Institute for Genetic Engineering and Biotechnology: 245.
- P. Ralph (2013). "The Geography of Recent Genetic Ancestry across Europe". PLOS Biology. 11 (5): e105090. doi:10.1371/journal.pbio.1001555. PMC 3646727. PMID 23667324.
- Kushniarevich, A; Utevska, O; Chuhryaeva, M; et al. (2015). "Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data". PLOS ONE. 10 (9): e0135820. Bibcode:2015PLoSO..1035820K. doi:10.1371/journal.pone.0135820. PMC 4558026. PMID 26332464.
- Kovacevic, Lejla; Tambets, Kristiina; Ilumäe, Anne-Mai; Kushniarevich, Alena; Yunusbayev, Bayazit; Solnik, Anu; Bego, Tamer; Primorac, Dragan; Skaro, Vedrana (2014-08-22). "Standing at the Gateway to Europe - The Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers". PLOS ONE. 9 (8): e105090. Bibcode:2014PLoSO...9j5090K. doi:10.1371/journal.pone.0105090. ISSN 1932-6203. PMC 4141785. PMID 25148043.
- Novembre, J; Johnson, T; Bryc, K; et al. (2008). "(November 2008), "Genes mirror geography within Europe". Nature. 456 (7218): 98–101. Bibcode:2008Natur.456...98N. doi:10.1038/nature07331. PMC 2735096. PMID 18758442.
- Marjanovic, D; Fornarino, S; Montagna, S; et al. (November 2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Ann. Hum. Genet. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Rebała, K.; et al. (2007). "Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin". J Hum Genet. 52 (5): 406–14. doi:10.1007/s10038-007-0125-6. PMID 17364156.
- "I-P37 YTree v6.07.08". YFull.com. 10 November 2018. Retrieved 11 November 2018.
- "I2a Y-Haplogroup - Results: I2a2a-Dinaric". Family Tree DNA. Retrieved 11 November 2018.
Ken Nordtvedt has split I2a2-M423-Dinaric into Din-N and Din-S. Din-N is older than Din-S. N=north of the Danube and S=south of the Danube River ... May 8, 2007: Dinaric I1b1 and DYS 448. DYS448 19 for S and 20 for N.
- Bernie Cullen (23 September 2016). "The Dinaric-South cluster is defined by the SNP PH908". i2aproject.blogspot.com. Blogger. Retrieved 11 November 2018.
- "Y-DNA Haplogroup I and its Subclades - 2018". ISOGG. 1 November 2018. Retrieved 11 November 2018.
- "I-PH908 YTree v6.07.08". YFull.com. 10 November 2018. Retrieved 11 November 2018.
- Utevska 2017, p. 219–226, 302.
- "I-PH908 - YFull YTree Info". www.yfull.com. Retrieved 2020-02-08.
- Neparáczki, Endre; et al. (2019). "Y-chromosome haplogroups from Hun, Avar and conquering Hungarian period nomadic people of the Carpathian Basin". Scientific Reports. Nature Research. 9 (16569): 16569. doi:10.1038/s41598-019-53105-5. PMC 6851379. PMID 31719606.
- Mathieson, Iain (February 21, 2018). "The Genomic History of Southeastern Europe". Nature. 555 (7695): 197–203. doi:10.1038/nature25778. PMC 6091220. PMID 29466330.
- Lipson, Mark (2017). "Parallel ancient genomic transects reveal complex population history of early European farmers". Nature. 551: 368–372. doi:10.1038/nature24476.
- Fóthi, E.; Gonzalez, A.; Fehér, T.; et al. (2020), "Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes", Archaeological and Anthropological Sciences, 12 (1), doi:10.1007/s12520-019-00996-0
- Peričić, M; Barać Lauc, L; Martinović, I; et al. (2005). "High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations" (PDF). Molecular Biology and Evolution. 22 (10): 1964–75. doi:10.1093/molbev/msi185. PMID 15944443.
- Dogan, S; Babic, N; Gurkan, C; et al. (2016). "Y-chromosomal haplogroup distribution in the Tuzla Canton of Bosnia and Herzegovina: A concordance study using four different in silico assignment algorithms based on Y-STR data". Journal HOMO of Comparative Human Biology. 67 (6): 471–483. doi:10.1016/j.jchb.2016.10.003. PMID 27908490.
- Marjanović, D; Fornarino, S; Montagna, S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Annals of Human Genetics. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Marjanović, D; Fornarino, S; Montagna, S; et al. (2005). "The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups". Annals of Human Genetics. 69 (Pt 6): 757–63. doi:10.1111/j.1529-8817.2005.00190.x. PMID 16266413.
- Doğan, S; Ašić, A; Doğan, G; et al. (2017). "Y chromosome haplogroups in the BosnianHerzegovinian population based on 23 Y-STR loci" (PDF). Hum. Biol. 88 (Pt 3): 201–209. doi:10.13110/humanbiology.88.3.0201. PMID 28828942.
- Peričić, M; Barać Lauc, L; Martinović, I; et al. (2005). "High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations" (PDF). Molecular Biology and Evolution. 22 (10): 1964–75. doi:10.1093/molbev/msi185. PMID 15944443.
- Dogan, S; Babic, N; Gurkan, C; et al. (2016). "Y-chromosomal haplogroup distribution in the Tuzla Canton of Bosnia and Herzegovina: A concordance study using four different in silico assignment algorithms based on Y-STR data". Journal HOMO of Comparative Human Biology. 67 (6): 471–483. doi:10.1016/j.jchb.2016.10.003. PMID 27908490.
- Malyarchuk, B.A.; Grzybowski, T.; Derenko, M. V.; et al. (2003). "Mitochondrial DNA Variability in Bosnians and Slovenes". Annals of Human Genetics. 67 (5): 412–425. doi:10.1046/j.1469-1809.2003.00042.x. PMID 12940915.
- Ahmić, A.; Hadžiselimović, R.; Silajdžić, E.; Mujkić, I.; Pojskić, N.; et al. (June 2019). "MtDNA variations in three main ethnic groups in Tuzla Canton of Bosnia and Herzegovina". Genetics & Applications. 3 (1): 13–23. doi:10.31383/ga.vol3iss1pp14-23.
- The SNP I-P37 itself formed approximately 20 thousand YBP and had TMRCA 18 thousand YBP according to YFull,[12] being too old and widespread as an SNP for argumentation of autochthony as well the old research used outdated nomenclature. According to "I-P37 (I2a)" project at Family Tree DNA, the divergence at STR marker DYS448 20 > 19 is reported since 2007,[13] while the SNP which defines the STR Dinaric-South cluster, I-PH908, is reported since 2016.[14] The SNP I-PH908 at ISOGG phylogenetic tree is named as I2a1a2b1a1a1c,[15] while formed and had TMRCA approximately 1,800 YBP according to YFull.[16]