Chromalveolata
Chromalveolata was a eukaryote supergroup present in a major classification of 2005, then regarded as one of the six major groups within the eukaryotes.[3] It was a refinement of the kingdom Chromista, first proposed by Thomas Cavalier-Smith in 1981. Chromalveolata was proposed to represent the organisms descended from a single secondary endosymbiosis involving a red alga and a bikont.[4] The plastids in these organisms are those that contain chlorophyll c.
| Chromalveolata | |
|---|---|
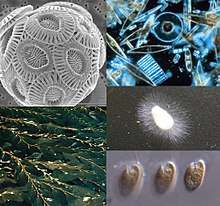 | |
| Clockwise from top-left: a haptophyte, some diatoms, a water mold, a cryptomonad, and Macrocystis, a phaeophyte | |
| Scientific classification | |
| (unranked): | Bikonta |
| (unranked): | Chromalveolata Adl et al., 2005 (not a monophyletic group)[1][2] |
| Phyla | |
However, the monophyly of the Chromalveolata has been rejected. Thus, two papers published in 2008 have phylogenetic trees in which the chromalveolates are split up,[5][6] and recent studies continue to support this view.[7][8]
Groups and classification
Historically, many chromalveolates were considered plants, because of their cell walls, photosynthetic ability, and in some cases their morphological resemblance to the land plants (Embryophyta). However, when the five-kingdom system (proposed in 1969) took prevalence over the animal–plant dichotomy, most of what we now call chromalveolates were put into the kingdom Protista, but the water molds and slime nets were put into the kingdom Fungi, while the brown algae stayed in the plant kingdom. These various organisms were later grouped together and given the name Chromalveolata by Cavalier-Smith. He believed them to be a monophyletic group, but this is not the case.[9]
In 2005, in a classification reflecting the consensus at the time, the Chromalveolata were regarded as one of the six major clades of eukaryotes.[3] Although not given a formal taxonomic status in this classification, elsewhere the group had been treated as a Kingdom. The Chromalveolata were divided into four major subgroups:
- Cryptophyta
- Haptophyta
- Stramenopiles (or Heterokontophyta)
- Alveolata
Other groups that may be included within, or related to, chromalveolates, are:
Though several groups, such as the ciliates and the water molds, have lost the ability to photosynthesize, most are autotrophic. All photosynthetic chromalveolates use chlorophylls a and c, and many use accessory pigments. Chromalveolates share similar glyceraldehyde 3-phosphate dehydrogenase proteins.[11]
However, as early as 2005, doubts were being expressed as to whether Chromalveolata was monophyletic,[9] and a review in 2006 noted the lack of evidence for several of the supposed six major eukaryote groups, including the Chromalveolata.[12] In 2012, consensus emerged that the group is not monophyletic. The four original subgroups fall into at least two categories: one comprises the Stramenopiles and the Alveolata, to which the Rhizaria are now usually added to form the SAR group; the other comprises the Cryptophyta and the Haptophyta.[5][6] A 2010 paper splits the Cryptophyta and Haptophyta; the former are a sister group to the SAR group, the latter cluster with the Archaeplastida (plants in the broad sense). The katablepharids are closely related to the cryptophytes and the telonemids and centrohelids may be related to the haptophytes.[7]
A variety of names have been used for different combinations of the groups formerly thought to make up the Chromalveolata.
- Halvaria Analyses in 2007 and 2008 agreed that the Stramenopiles and the Alveolata were related, forming a reduced chromalveolate clade, called Halvaria.[5][6][13]
- SAR group The Rhizaria, which were originally not considered to be chromalveolates, belong with the Stramenopiles and Alveolata in many analyses, forming the SAR group, i.e. Halvaria plus Rhizaria.[13][14]
- Hacrobia The other two groups originally included in Chromalveolata, the Haptophyta and the Cryptophyta, were related in some analyses,[5][6] forming a clade which has been called Hacrobia. Alternatively, the Hacrobia appeared to be more closely related to the Archaeplastida (plants in the very broad sense), being a sister group in one analysis,[5] and actually nested inside this group in another.[6] (Earlier, Cavalier-Smith had suggested a clade called Corticata for the grouping of all the chromalveolates and the Archaeplastida.) More recently, as noted above, Hacrobia has been split, with the Haptophyta being sister to the SAR group and the Cryptophyta instead related to the Archaeplastida.[7]
Morphology
Chromalveolates, unlike other groups with multicellular representatives, do not have very many common morphological characteristics. Each major subgroup has certain unique features, including the alveoli of the Alveolata, the haptonema of the Haptophyta, the ejectisome of the Cryptophyta, and the two different flagella of the Heterokontophyta. However, none of these features are present in all of the groups.
The only common chromalveolate features are these:
- The shared origin of chloroplasts, as mentioned above
- Presence of cellulose in most cell walls
Since this is such a diverse group, it is difficult to summarize shared chromalveolate characteristics.
Ecological role

Many chromalveolates affect our ecosystem in enormous ways.
Some of these organisms can be very harmful. Dinoflagellates produce red tides, which can devastate fish populations and intoxicate oyster harvests. Apicomplexans are some of the most successful specific parasites to animals (including the genus Plasmodium, the malaria parasites). Water molds cause several plant diseases - it was the water mold Phytophthora infestans that caused the Irish potato blight that led to the Great Irish Famine.
However, many others are vital members of our ecosystem. Diatoms are one of the major photosynthetic producers, and as such produce much of the oxygen that we breathe, and also take in much of the carbon dioxide from the atmosphere. Brown algae, most specifically kelps, create underwater "forest" habitats for many marine creatures, and provide a large portion of the diet of coastal communities.
Chromalveolates also provide many products that we use. The algin in brown algae is used as a food thickener, most famously in ice cream. The siliceous shells of diatoms have many uses, such as in reflective paint, in toothpaste, or as a filter, in what is known as diatomaceous earth.
Chromalveolata viruses
Like other organisms, chromalveolata have viruses. In the case of Emiliania huxleyi (a common algal bloom chromalveolate), a virus believed to be specific to it causes mass death and the end of the bloom.[15]
See also
References
- Katz, Laura A.; Grant, Jessica R. (2015-05-01). "Taxon-Rich Phylogenomic Analyses Resolve the Eukaryotic Tree of Life and Reveal the Power of Subsampling by Sites". Systematic Biology. 64 (3): 406–415. doi:10.1093/sysbio/syu126. ISSN 1063-5157. PMID 25540455.
- Cavalier-Smith, Thomas; Chao, Ema E.; Lewis, Rhodri (2015-12-01). "Multiple origins of Heliozoa from flagellate ancestors: New cryptist subphylum Corbihelia, superclass Corbistoma, and monophyly of Haptista, Cryptista, Hacrobia and Chromista". Molecular Phylogenetics and Evolution. 93: 331–362. doi:10.1016/j.ympev.2015.07.004. PMID 26234272.
- Adl, Sina M.; et al. (2005), "The New Higher Level Classification of Eukaryotes with Emphasis on the Taxonomy of Protists", Journal of Eukaryotic Microbiology, 52 (5): 399–451, doi:10.1111/j.1550-7408.2005.00053.x, PMID 16248873, S2CID 8060916
- Keeling PJ (2009). "Chromalveolates and the evolution of plastids by secondary endosymbiosis". J. Eukaryot. Microbiol. 56 (1): 1–8. doi:10.1111/j.1550-7408.2008.00371.x. PMID 19335769. S2CID 34259721.
- Burki, Fabien; Shalchian-Tabrizi, Kamran & Pawlowski, Jan (2008). "Phylogenomics reveals a new 'megagroup' including most photosynthetic eukaryotes". Biology Letters. 4 (4): 366–369. doi:10.1098/rsbl.2008.0224. PMC 2610160. PMID 18522922.
- Kim, E; Graham, LE (Jul 2008). Redfield, Rosemary Jeanne (ed.). "EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata". PLOS ONE. 3 (7): e2621. Bibcode:2008PLoSO...3.2621K. doi:10.1371/journal.pone.0002621. PMC 2440802. PMID 18612431.
- Burki, F.; Okamoto, N.; Pombert, J.F. & Keeling, P.J. (2012). "The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins". Proc. Biol. Sci. 279 (1736): 2246–2254. doi:10.1098/rspb.2011.2301. PMC 3321700. PMID 22298847.
- Burki, Fabien; Kaplan, Maia; Tikhonenkov, Denis V.; Zlatogursky, Vasily; Minh, Bui Quang; Radaykina, Liudmila V.; Smirnov, Alexey; Mylnikov, Alexander P.; Keeling, Patrick J. (2016-01-27). "Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista". Proc. R. Soc. B. 283 (1823): 20152802. doi:10.1098/rspb.2015.2802. ISSN 0962-8452. PMC 4795036. PMID 26817772.
- Harper, J. T., Waanders, E. & Keeling, P. J. 2005. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int. J. System. Evol. Microbiol., 55, 487-496. "Archived copy" (PDF). Archived from the original (PDF) on 2008-12-17. Retrieved 2010-04-26.CS1 maint: archived copy as title (link)
- Shalchian-Tabrizi K, Eikrem W, Klaveness D, Vaulot D, Minge M, Le Gall F, Romari K, Throndsen J, Botnen A, Massana R, Thomsen H, Jakobsen K (2006). "Telonemia, a new protist phylum with affinity to chromist lineages". Proc Biol Sci. 273 (1595): 1833–42. doi:10.1098/rspb.2006.3515. PMC 1634789. PMID 16790418.
- Takishita K, Yamaguchi H, Maruyama T, Inagaki Y (2009). Zhang B (ed.). "A hypothesis for the evolution of nuclear-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase genes in "chromalveolate" members". PLOS ONE. 4 (3): e4737. Bibcode:2009PLoSO...4.4737T. doi:10.1371/journal.pone.0004737. PMC 2649427. PMID 19270733.
- Laura Wegener Parfrey; Erika Barbero; Elyse Lasser; Micah Dunthorn; Debashish Bhattacharya; David J Patterson; Laura A Katz (December 2006). "Evaluating Support for the Current Classification of Eukaryotic Diversity". PLOS Genet. 2 (12): e220. doi:10.1371/journal.pgen.0020220. PMC 1713255. PMID 17194223.
- Fabien Burki; Kamran Shalchian-Tabrizi; Marianne Minge; Åsmund Skjæveland; Sergey I. Nikolaev; Kjetill S. Jakobsen; Jan Pawlowski (2007). "Phylogenomics Reshuffles the Eukaryotic Supergroups". PLOS ONE. 2 (8): e790. Bibcode:2007PLoSO...2..790B. doi:10.1371/journal.pone.0000790. PMC 1949142. PMID 17726520.
- Hampl V, Hug L, Leigh JW, et al. (March 2009). "Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups"". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3859–64. Bibcode:2009PNAS..106.3859H. doi:10.1073/pnas.0807880106. PMC 2656170. PMID 19237557.
- Madhusoodanan, Jyoti (August 24, 2014). "Viral demise of an algal bloom:Marine viruses may be key players in the death of massive algal blooms that emerge in the ocean, a study shows". TheScientist.
External links
| Wikispecies has information related to Chromalveolata |
| Wikimedia Commons has media related to Chromalveolata. |
