Lanosterol 14 alpha-demethylase
Lanosterol 14α-demethylase (CYP51A1) is a cytochrome P450 enzyme that is involved in the conversion of lanosterol to 4,4-dimethylcholesta-8(9),14,24-trien-3β-ol.[4] The cytochrome P450 isoenzymes are a conserved group of proteins that serve as key players in the metabolism of organic substances and the biosynthesis of important steroids, lipids, and vitamins in eukaryotes.[5] As a member of this family, lanosterol 14α-demethylase is responsible for an essential step in the biosynthesis of sterols. In particular, this protein catalyzes the removal of the C-14α-methyl group from lanosterol.[5] This demethylation step is regarded as the initial checkpoint in the transformation of lanosterol to other sterols that are widely used within the cell.[5]
| Cytochrome P450, Family 51, Subfamily A, Polypeptide 1 | |
|---|---|
| Identifiers | |
| Symbol | CYP51A1 |
| Alt. symbols | CYP51, P45014DM |
| NCBI gene | 1595 |
| HGNC | 2649 |
| OMIM | 601637 |
| RefSeq | NM_000786 |
| UniProt | Q16850 |
| Other data | |
| EC number | 1.14.14.154 |
| Locus | Chr. 7 q21.2-21.3 |
Evolution
The structural and functional properties of the cytochrome P450 superfamily have been subject to extensive diversification over the course of evolution.[6] Recent estimates indicate that there are currently 10 classes and 267 families of CYP proteins.[7] It is believed that 14α-demethylase or CYP51 diverged early in the cytochrome's evolutionary history and has preserved its function ever since; namely, the removal of the 14α-methyl group from sterol substrates.[6]
Although CYP51's mode of action has been well conserved, the protein's sequence varies considerably between biological kingdoms.[8] CYP51 sequence comparisons between kingdoms reveal only a 22-30% similarity in amino acid composition.[9]
Structure
.png)
Although the structure of 14α-demethylase may vary substantially from one organism to the next, sequence alignment analysis reveals that there are six regions in the protein that are highly conserved in eukaryotes.[9] These include residues in the B' helix, B'/C loop, C helix, I helix, K/β1-4 loop, and β-strand 1-4 that are responsible for forming the surface of the substrate binding cavity.[6] Homology modeling reveals that substrates migrate from the surface of the protein to the enzyme's buried active site through a channel that is formed in part by the A' alpha helix and the β4 loop.[10][11] Finally, the active site contains a heme prosthetic group in which the iron is tethered to a thiolate ligand on a conserved cysteine residue.[9] This group also binds diatomic oxygen at the sixth coordination site, which is eventually incorporated onto the substrate.[9]
Mechanism
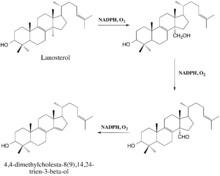
The enzyme-catalyzed demethylation of lanosterol is believed to occur in three steps, each of which requires one molecule of diatomic oxygen and one molecule of NADPH (or some other reducing equivalent).[12] During the first two steps, the 14α-methyl group undergoes typical cytochrome monooxygenation in which one oxygen atom is incorporated by the substrate and the other is reduced to water, resulting in the sterol's conversion to a carboxyalcohol and then a carboxyaldehyde.[9] The aldehyde then departs as formic acid and a double bond is simultaneously introduced to yield the demethylated product.[9]
See also
- Steroidogenic enzyme
- Azole antifungals
References
- GRCh38: Ensembl release 89: ENSG00000001630 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Metabocard for 4,4-Dimethylcholesta-8,14,24-trienol (HMDB01023)". Human Metabolome Database. February 2014.
- Lepesheva GI, Waterman MR (March 2007). "Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms". Biochimica et Biophysica Acta (BBA) - General Subjects. 1770 (3): 467–77. doi:10.1016/j.bbagen.2006.07.018. PMC 2324071. PMID 16963187.
- Becher R, Wirsel SG (August 2012). "Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens". Applied Microbiology and Biotechnology. 95 (4): 825–40. doi:10.1007/s00253-012-4195-9. PMID 22684327.
- Hannemann F, Bichet A, Ewen KM, Bernhardt R (March 2007). "Cytochrome P450 systems--biological variations of electron transport chains". Biochimica et Biophysica Acta (BBA) - General Subjects. 1770 (3): 330–44. doi:10.1016/j.bbagen.2006.07.017. PMID 16978787.
- Lepesheva GI, Waterman MR (February 2004). "CYP51--the omnipotent P450". Molecular and Cellular Endocrinology. 215 (1–2): 165–70. doi:10.1016/j.mce.2003.11.016. PMID 15026190.
- Lepesheva GI, Waterman MR (January 2011). "Structural basis for conservation in the CYP51 family". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1814 (1): 88–93. doi:10.1016/j.bbapap.2010.06.006. PMC 2962772. PMID 20547249.
- Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI (July 2011). "Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14alpha-demethylase (CYP51) from Leishmania infantum". The Journal of Biological Chemistry. 286 (30): 26838–48. doi:10.1074/jbc.M111.237099. PMC 3143644. PMID 21632531.
- Podust LM, von Kries JP, Eddine AN, Kim Y, Yermalitskaya LV, Kuehne R, et al. (November 2007). "Small-molecule scaffolds for CYP51 inhibitors identified by high-throughput screening and defined by X-ray crystallography". Antimicrobial Agents and Chemotherapy. 51 (11): 3915–23. doi:10.1128/AAC.00311-07. PMC 2151439. PMID 17846131.
- Vanden Bossche H, Koymans L (1998). "Cytochromes P450 in fungi". Mycoses. 41 Suppl 1: 32–8. doi:10.1111/j.1439-0507.1998.tb00581.x. PMID 9717384.
Further reading
- Bak S, Kahn RA, Olsen CE, Halkier BA (February 1997). "Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals". The Plant Journal. 11 (2): 191–201. doi:10.1046/j.1365-313X.1997.11020191.x. PMID 9076987.
- Aoyama Y, Yoshida Y (August 1991). "Different substrate specificities of lanosterol 14a-demethylase (P-45014DM) of Saccharomyces cerevisiae and rat liver for 24-methylene-24,25-dihydrolanosterol and 24,25-dihydrolanosterol". Biochemical and Biophysical Research Communications. 178 (3): 1064–71. doi:10.1016/0006-291X(91)91000-3. PMID 1872829.
- Aoyama Y, Yoshida Y (March 1992). "The 4 beta-methyl group of substrate does not affect the activity of lanosterol 14 alpha-demethylase (P-450(14)DM) of yeast: difference between the substrate recognition by yeast and plant sterol 14 alpha-demethylases". Biochemical and Biophysical Research Communications. 183 (3): 1266–72. doi:10.1016/S0006-291X(05)80327-4. PMID 1567403.
- Alexander K, Akhtar M, Boar RB, McGhie JF, Barton DH (1972). "The removal of the 32-carbon atom as formic acid in cholesterol biosynthesis". Journal of the Chemical Society, Chemical Communications (7): 383. doi:10.1039/C39720000383.
External links
- cytochrome+P-450+CYP51 at the US National Library of Medicine Medical Subject Headings (MeSH)