Virology
Virology is the study of viruses – submicroscopic, parasitic particles of genetic material contained in a protein coat[1][2] – and virus-like agents. It focuses on the following aspects of viruses: their structure, classification and evolution, their ways to infect and exploit host cells for reproduction, their interaction with host organism physiology and immunity, the diseases they cause, the techniques to isolate and culture them, and their use in research and therapy. Virology is a subfield of microbiology.
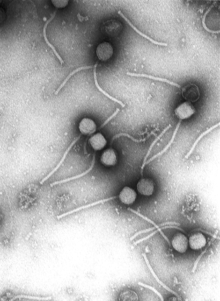
Virus structure and classification
A major branch of virology is virus classification. Viruses can be classified according to the host cell they infect: animal viruses, plant viruses, fungal viruses, and bacteriophages (viruses infecting bacterium, which include the most complex viruses). Another classification uses the geometrical shape of their capsid (often a helix or an icosahedron) or the virus's structure (e.g. presence or absence of a lipid envelope).[3] Viruses range in size from about 30 nm to about 450 nm, which means that most of them cannot be seen with light microscopes. The shape and structure of viruses has been studied by electron microscopy, NMR spectroscopy, and X-ray crystallography.
The most useful and most widely used classification system distinguishes viruses according to the type of nucleic acid they use as genetic material and the viral replication method they employ to coax host cells into producing more viruses:
- DNA viruses (divided into double-stranded DNA viruses and single-stranded DNA viruses),
- RNA viruses (divided into positive-sense single-stranded RNA viruses, negative-sense single-stranded RNA viruses and the much less common double-stranded RNA viruses),
- reverse transcribing viruses (double-stranded reverse-transcribing DNA viruses and single-stranded reverse-transcribing RNA viruses including retroviruses).
The latest report by the International Committee on Taxonomy of Viruses (2005) lists 5,450 viruses, organized in over 2,000 species, 287 genera, 73 families and 3 orders.
Virologists also study subviral particles, infectious entities notably smaller and simpler than viruses:
- viroids (naked circular RNA molecules infecting plants),
- satellites (nucleic acid molecules with or without a capsid that require a helper virus for infection and reproduction), and
- prions (proteins that can exist in a pathological conformation that induces other prion molecules to assume that same conformation).[4]
Taxa in virology are not necessarily monophyletic, as the evolutionary relationships of the various virus groups remain unclear. Three hypotheses regarding their origin exist:
- Viruses arose from non-living matter, separately from yet in parallel to cells, perhaps in the form of self-replicating RNA ribozymes similar to viroids.
- Viruses arose by genome reduction from earlier, more competent cellular life forms that became parasites to host cells and subsequently lost most of their functionality; examples of such tiny parasitic prokaryotes are Mycoplasma and Nanoarchaea.
- Viruses arose from mobile genetic elements of cells (such as transposons, retrotransposons or plasmids) that became encapsulated in protein capsids, acquired the ability to "break free" from the host cell and infect other cells.
Of particular interest here is mimivirus, a giant virus that infects amoebae and encodes much of the molecular machinery traditionally associated with bacteria. Two possibilities are that it is a simplified version of a parasitic prokaryote or it originated as a simpler virus that acquired genes from its host.
The evolution of viruses, which often occurs in concert with the evolution of their hosts, is studied in the field of viral evolution.
While viruses reproduce and evolve, they do not engage in metabolism, do not move, and depend on a host cell for reproduction. The often-debated question of whether they are alive or not is a matter of definition that does not affect the biological reality of viruses.
Viral diseases and host defenses
One main motivation for the study of viruses is the fact that they cause many important infectious diseases, among them the common cold, influenza, rabies, measles, many forms of diarrhea, hepatitis, Dengue fever, yellow fever, polio, smallpox and AIDS.[5] Herpes simplex causes cold sores and genital herpes and is under investigation as a possible factor in Alzheimer's.[6]
Some viruses, known as oncoviruses, contribute to the development of certain forms of cancer. The best-studied example is the association between Human papillomavirus and cervical cancer: almost all cases of cervical cancer are caused by certain strains of this sexually transmitted virus. Another example is the association of infection with hepatitis B and hepatitis C viruses and liver cancer.
Some subviral particles also cause disease: the transmissible spongiform encephalopathies, which include Kuru, Creutzfeldt–Jakob disease and bovine spongiform encephalopathy ("mad cow disease"), are caused by prions,[7] hepatitis D is due to a satellite virus.
The study of the manner in which viruses cause disease is viral pathogenesis. The degree to which a virus causes disease is its virulence.
When the immune system of a vertebrate encounters a virus, it may produce specific antibodies which bind to the virus and neutralize its infectivity or mark it for destruction. Antibody presence in blood serum is often used to determine whether a person has been exposed to a given virus in the past, with tests such as ELISA. Vaccinations protect against viral diseases, in part, by eliciting the production of antibodies. Monoclonal antibodies, specific to the virus, are also used for detection, as in fluorescence microscopy.
The second defense of vertebrates against viruses, cell-mediated immunity, involves immune cells known as T cells: the body's cells constantly display short fragments of their proteins on the cell's surface, and if a T cell recognizes a suspicious viral fragment there, the host cell is destroyed and the virus-specific T-cells proliferate. This mechanism is jump-started by certain vaccinations.
RNA interference, an important cellular mechanism found in plants, animals and many other eukaryotes, most likely evolved as a defense against viruses. An elaborate machinery of interacting enzymes detects double-stranded RNA molecules (which occur as part of the life cycle of many viruses) and then proceeds to destroy all single-stranded versions of those detected RNA molecules.
Every lethal viral disease presents a paradox: killing its host is obviously of no benefit to the virus, so how and why did it evolve to do so? Today it is believed that most viruses are relatively benign in their natural hosts; some viral infection might even be beneficial to the host.[8] The lethal viral diseases are believed to have resulted from an "accidental" jump of the virus from a species in which it is benign to a new one that is not accustomed to it (see zoonosis). For example, viruses that cause serious influenza in humans probably have pigs or birds as their natural host, and HIV is thought to derive from the benign non-human primate virus SIV.
While it has been possible to prevent (certain) viral diseases by vaccination for a long time, the development of antiviral drugs to treat viral diseases is a comparatively recent development. The first such drug was interferon, a substance that is naturally produced when an infection is detected and stimulates other parts of the immune system.
Molecular biology research and viral therapy
Bacteriophages, the viruses which infect bacteria, can be relatively easily grown as viral plaques on bacterial cultures. Bacteriophages occasionally move genetic material from one bacterial cell to another in a process known as transduction,[9] and this horizontal gene transfer is one reason why they served as a major research tool in the early development of molecular biology. The genetic code, the function of ribozymes, the first recombinant DNA and early genetic libraries were all arrived at using bacteriophages. Certain genetic elements derived from viruses, such as highly effective promoters, are commonly used in molecular biology research today.
Growing animal viruses outside of the living host animal is more difficult. Classically, fertilized chicken eggs have often been used, but cell cultures are increasingly employed for this purpose today.
Since some viruses that infect eukaryotes need to transport their genetic material into the host cell's nucleus, they are attractive tools for introducing new genes into the host (known as transformation or transfection). Modified retroviruses are often used for this purpose, as they integrate their genes into the host's chromosomes.
This approach of using viruses as gene vectors is being pursued in the gene therapy of genetic diseases. An obvious problem to be overcome in viral gene therapy is the rejection of the transforming virus by the immune system.
Phage therapy, the use of bacteriophages to combat bacterial diseases, was a popular research topic before the advent of antibiotics and has recently seen renewed interest.
Oncolytic viruses are viruses that preferably infect cancer cells. While early efforts to employ these viruses in the therapy of cancer failed, there have been reports in 2005 and 2006 of encouraging preliminary results.[10]
Sequencing of viruses
As most viruses are too small to be seen by a light microscope, sequencing is one of the main tools in virology to identify and study the virus. Traditional Sanger sequencing and next-generation sequencing (NGS) are used to sequence viruses in basic and clinical research, as well as for the diagnosis of emerging viral infections, molecular epidemiology of viral pathogens, and drug-resistance testing. There are more than 2.3 million unique viral sequences in GenBank.[11] Recently, NGS has surpassed traditional Sanger as the most popular approach for generating viral genomes.[11]
Other uses of viruses
A new application of genetically engineered viruses in nanotechnology was recently described; see the uses of viruses in material science and nanotechnology. For use in mapping neurons see the applications of pseudorabies in neuroscience.
History of virology


The word virus appeared in 1599 and originally meant "venom".[12]
A very early form of vaccination known as variolation was developed several thousand years ago in China. It involved the application of materials from smallpox sufferers in order to immunize others. In 1717 Lady Mary Wortley Montagu observed the practice in Istanbul and attempted to popularize it in Britain, but encountered considerable resistance. In 1796 Edward Jenner developed a much safer method, using cowpox to successfully immunize a young boy against smallpox, and this practice was widely adopted. Vaccinations against other viral diseases followed, including the successful rabies vaccination by Louis Pasteur in 1886. The nature of viruses, however, was not clear to these researchers.
In 1892, the Russian biologist Dmitry Ivanovsky used a Chamberland filter to try to isolate the bacteria that caused tobacco mosaic disease. His experiments showed that crushed leaf extracts from infected tobacco plants remained infectious after filtration. Ivanovsky reported a minuscule infectious agent or toxin, capable of passing the filter, may be being produced by a bacterium.[13][14][15]
In 1898 Martinus Beijerinck repeated Ivanovski's work but went further and passed the "filterable agent" from plant to plant, found the action undiminished, and concluded it infectious – replicating in the host – and thus not a mere toxin. He called it contagium vivum fluidum.[16] The question of whether the agent was a "living fluid" or a particle was however still open.
In 1903 it was suggested for the first time that transduction by viruses might cause cancer. In 1908 Bang and Ellerman showed that a filterable virus could transmit chicken leukemia, data largely ignored till the 1930s when leukemia became regarded as cancerous.[17] In 1911 Peyton Rous reported the transmission of chicken sarcoma, a solid tumor, with a virus, and thus Rous became "father of tumor virology".[17] The virus was later called Rous sarcoma virus 1 and understood to be a retrovirus. Several other cancer-causing retroviruses have since been described.
The existence of viruses that infect bacteria (bacteriophages) was first recognized by Frederick Twort in 1911, and, independently, by Félix d'Herelle in 1917. As bacteria could be grown easily in culture, this led to an explosion of virology research.
The cause of the devastating Spanish flu pandemic of 1918 was initially unclear. In late 1918, French scientists showed that a "filter-passing virus" could transmit the disease to people and animals, fulfilling Koch's postulates.[18]
In 1926 it was shown that scarlet fever is caused by a bacterium that is infected by a certain bacteriophage.
While plant viruses and bacteriophages can be grown comparatively easily, animal viruses normally require a living host animal, which complicates their study immensely. In 1931 it was shown that influenza virus could be grown in fertilized chicken eggs, a method that is still used today to produce vaccines. In 1937, Max Theiler managed to grow the yellow fever virus in chicken eggs and produced a vaccine from an attenuated virus strain; this vaccine saved millions of lives and is still being used today.
Max Delbrück, an important investigator in the area of bacteriophages, described the basic "life cycle" of a virus in 1937: rather than "growing", a virus particle is assembled from its constituent pieces in one step; eventually it leaves the host cell to infect other cells. The Hershey–Chase experiment in 1952 showed that only DNA and not protein enters a bacterial cell upon infection with bacteriophage T2. Transduction of bacteria by bacteriophages was first described in the same year.
In 1949 John F. Enders, Thomas Weller and Frederick Robbins reported growth of poliovirus in cultured human embryonal cells, the first significant example of an animal virus grown outside of animals or chicken eggs. This work aided Jonas Salk in deriving a polio vaccine from deactivated polio viruses; this vaccine was shown to be effective in 1955.
The first virus that could be crystalized and whose structure could, therefore, be elucidated in detail was tobacco mosaic virus (TMV), the virus that had been studied earlier by Ivanovski and Beijerink. In 1935, Wendell Stanley achieved its crystallization for electron microscopy and showed that it remains active even after crystallization. Clear X-ray diffraction pictures of the crystallized virus were obtained by Bernal and Fankuchen in 1941. Based on such pictures, Rosalind Franklin proposed the full structure of the tobacco mosaic virus in 1955. Also in 1955, Heinz Fraenkel-Conrat and Robley Williams showed that purified TMV RNA and its capsid (coat) protein can self-assemble into functional virions, suggesting that this assembly mechanism is also used within the host cell, as Delbrück had proposed earlier.
In 1963, the Hepatitis B virus was discovered by Baruch Blumberg who went on to develop a hepatitis B vaccine.
In 1965, Howard Temin described the first retrovirus: a virus whose RNA genome was reverse transcribed into complementary DNA (cDNA), then integrated into the host's genome and expressed from that template. The viral enzyme reverse transcriptase, which along with integrase is a distinguishing trait of retroviruses, was first described in 1970, independently by Howard Temin and David Baltimore. The first retrovirus infecting humans was identified by Robert Gallo in 1974. Later it was found that reverse transcriptase is not specific to retroviruses; retrotransposons which code for reverse transcriptase are abundant in the genomes of all eukaryotes. About 10–40% of the human genome derives from such retrotransposons.
In 1975 the functioning of oncoviruses was clarified considerably. Until that time, it was thought that these viruses carried certain genes called oncogenes which, when inserted into the host's genome, would cause cancer. Michael Bishop and Harold Varmus showed that the oncogene of Rous sarcoma virus is in fact not specific to the virus but is contained in the genome of healthy animals of many species. The oncovirus can switch this pre-existing benign proto-oncogene on, turning it into a true oncogene that causes cancer.
1976 saw the first recorded outbreak of Ebola virus disease, a highly lethal virally transmitted disease.
In 1977, Frederick Sanger achieved the first complete sequencing of the genome of any organism, the bacteriophage Phi X 174. In the same year, Richard Roberts and Phillip Sharp independently showed that the genes of adenovirus contain introns and therefore require gene splicing. It was later realized that almost all genes of eukaryotes have introns as well.
A worldwide vaccination campaign led by the UN World Health Organization resulted in the eradication of smallpox in 1979.
In 1982, Stanley Prusiner discovered prions and showed that they cause scrapie.
The first cases of AIDS were reported in 1981, and HIV, the retrovirus causing it, was identified in 1983 by Luc Montagnier, Françoise Barré-Sinoussi and Robert Gallo.[19][20][21] Tests detecting HIV infection by detecting the presence of HIV antibody were developed. Subsequent tremendous research efforts turned HIV into the best studied virus. Human Herpes Virus 8, the cause of Kaposi's sarcoma which is often seen in AIDS patients, was identified in 1994. Several antiretroviral drugs were developed in the late 1990s, decreasing AIDS mortality dramatically in developed countries. Treatment that exists for HIV includes a multitude of different drugs collectively termed Highly Active Antiretroviral Therapy (HAART). HAART attacks many different aspects of the HIV virus, effectively reducing its effects below the limit of detection. However, when the administration of HAART is discontinued, HIV will bounce back. This is because HAART does not attack latently infected HIV cells, which can reactivate.[22]
The Hepatitis C virus was identified using novel molecular cloning techniques in 1987, leading to screening tests that dramatically reduced the incidence of post-transfusion hepatitis.[23]
The first attempts at gene therapy involving viral vectors began in the early 1980s, when retroviruses were developed that could insert a foreign gene into the host's genome. They contained the foreign gene but did not contain the viral genome and therefore could not reproduce. Tests in mice were followed by tests in humans, beginning in 1989. The first human studies attempted to correct the genetic disease severe combined immunodeficiency (SCID), but clinical success was limited. In the period from 1990 to 1995, gene therapy was tried on several other diseases and with different viral vectors, but it became clear that the initially high expectations were overstated. In 1999 a further setback occurred when 18-year-old Jesse Gelsinger died in a gene therapy trial. He suffered a severe immune response after having received an adenovirus vector. Success in the gene therapy of two cases of X-linked SCID was reported in 2000.[24]
In 2002 it was reported that poliovirus had been synthetically assembled in the laboratory, representing the first synthetic organism. Assembling the 7741-base genome from scratch, starting with the virus's published RNA sequence, took about two years. In 2003 a faster method was shown to assemble the 5386-base genome of the bacteriophage Phi X 174 in two weeks.
The giant mimivirus, in some sense an intermediate between tiny prokaryotes and ordinary viruses, was described in 2003 and sequenced in 2004.
The strain of Influenza A virus subtype H1N1 that killed up to 50 million people during the Spanish flu pandemic in 1918 was reconstructed in 2005. Sequence information was pieced together from preserved tissue samples of flu victims; viable virus was then synthesized from this sequence.[25] The 2009 flu pandemic involved another strain of Influenza A H1N1, commonly known as "swine flu".
By 1985, Harald zur Hausen had shown that two strains of Human papillomavirus (HPV) cause most cases of cervical cancer. Two vaccines protecting against these strains were released in 2006.
In 2006 and 2007 it was reported that introducing a small number of specific transcription factor genes into normal skin cells of mice or humans can turn these cells into pluripotent stem cells, known as induced pluripotent stem cells. The technique uses modified retroviruses to transform the cells; this is a potential problem for human therapy since these viruses integrate their genes at a random location in the host's genome, which can interrupt other genes and potentially causes cancer.[26]
In 2008, Sputnik virophage was described, the first known virophage: it uses the machinery of a helper virus to reproduce and inhibits reproduction of that helper virus. Sputnik reproduces in amoeba infected by mamavirus, a relative of the mimivirus mentioned above and the largest known virus to date.[27]
An endogenous retrovirus (ERV) is a viral element in the genome that was derived from a retrovirus whose genome has been incorporated into the germ-line genome of some organism and is therefore copied with each reproduction of that organism. It is estimated that about 9 percent of the human genome originates from ERVs. In 2015 it was shown that proteins from an ERV are actively expressed in 3-day-old human embryos and appear to play a role in embryonal development and protect embryos from infection by other viruses.[28]
Since the invention of Organ-on-a-chip in 2010s, the engineering approach has found application in the study of many diseases. The approach has also been introduced to virology and chip models are being developed. Examples include the invention of Influenza model by Donald E. Ingber group, the invention of Ebola virus disease model by Alireza Mashaghi group, and the invention of viral hepatitis model by Marcus Dorner group[29]. The organ chip approach will likely replace animal models for human virology.
See also
- Introduction to viruses
- Glossary of virology
- List of viruses
- Animal virology
- Category:Viral diseases
- List of viral diseases
- Wikipedia:WikiProject Viruses
References
- Crawford, Dorothy (2011). Viruses: A Very Short Introduction. New York: Oxford University Press. p. 4. ISBN 978-0199574858.
- Cann, Alan (2011). Principles of Molecular Virology (5 ed.). London: Academic Press. ISBN 978-0123849397.
- Themes, U. F. O. (2017-02-19). "6 Viruses—Basic Concepts". Basicmedical Key. Retrieved 2020-05-29.
- "Prion Diseases". CDC. Archived from the original on 2010-03-04. Retrieved 2016-03-25.
- Evans, Alfred (1982). Viral Infections of Humans. New York: Plenum Publishing Corporation. pp. xxv–xxxi. ISBN 0306406764.
- Lövheim H, Gilthorpe J, Adolfsson R, Nilsson LG, Elgh F (July 2014). "Reactivated herpes simplex infection increases the risk of Alzheimer's disease". Alzheimer's & Dementia. 11 (6): 593–99. doi:10.1016/j.jalz.2014.04.522. PMID 25043910.
- "1 Moved | Prion Diseases | CDC". Archived from the original on 2010-03-04. Retrieved 2017-09-17.
- Dimmock NJ, Easton AJ, Leppard K, Introduction to Modern Virology, (Oxford: Blackwell Publishers, 2007), ch 23 "Horizons in human virology", subch 23.3 "Subtle and insidious virus-host interactions", sec "Virus infections can give their host an evolutionary advantage", p. 432.
- Stanley Maloy. "Horizontal Gene Transfer". Retrieved 2016-03-25.
- Viruses: The new cancer hunters, IsraCast, 1 March 2006
- Castro, Christina; Marine, Rachel; Ramos, Edward; Ng, Terry Fei Fan (2019). "The effect of variant interference on de novo assembly for viral deep sequencing". bioRxiv: 815480. doi:10.1101/815480.
- "Virus", Merriam-Webster, Inc, 2011.
- Sussman, Max; Topley, W. W. C.; Wilson, Graham K.; Collier, L. H.; Balows, Albert (1998). Topley & Wilson's microbiology and microbial infections. London: Arnold. p. 3. ISBN 0-340-66316-2.
- Iwanowski, D. (1892). "Über die Mosaikkrankheit der Tabakspflanze". Bulletin Scientifique Publié Par l'Académie Impériale des Sciences de Saint-Pétersbourg / Nouvelle Serie III (in German and Russian). St. Petersburg. 35: 67–70. Translated into English in Johnson, J., Ed. (1942) Phytopathological classics (St. Paul, Minnesota: American Phytopathological Society) No. 7, pp. 27–30.
- Iwanowski, D. (1903). "Über die Mosaikkrankheit der Tabakspflanze". Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz (in German). 13: 1–41.
- Pennazio S (2007). "Genetics and virology: Two interdisciplinary branches of biology". Rivista di Biologia. 100 (1): 119–46. PMID 17592822.
- Van Epps HL (2005). "Peyton Rous: Father of the tumor virus" (PDF). Journal of Experimental Medicine. 201 (3): 320. doi:10.1084/jem.2013fta. PMC 2213042. PMID 15756727.
- The Medical and Scientific Conceptions of Influenza, Human Virology at Stanford
- Montagnier L (2002). "Historical essay. A History of HIV Discovery". Science. 298 (5599): 1727–28. doi:10.1126/science.1079027. PMID 12459575.
- Gallo RC (2002). "Historical essay. The Early Years of HIV/AIDS". Science. 298 (5599): 1728–30. doi:10.1126/science.1078050. PMID 12459576.
- Gallo RC, Montagnier L (2002). "Historical essay. Prospects for the Future". Science. 298 (5599): 1730–31. doi:10.1126/science.1079864. PMID 12459577.
- Hu, Wenhui; Kaminski, Rafal; Yang, Fan; Zhang, Yonggang; Cosentino, Laura; Li, Fang; Luo, Biao; Alvarez-Carbonell, David; Garcia-Mesa, Yoelvis (2014-08-05). "RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection". Proceedings of the National Academy of Sciences. 111 (31): 11461–66. Bibcode:2014PNAS..11111461H. doi:10.1073/pnas.1405186111. ISSN 0027-8424. PMC 4128125. PMID 25049410.
- 2000 Albert Lasker Award for Clinical Medical Research Archived October 28, 2007, at the Wayback Machine, The Lasker Foundation. Accessed 20 February 2008
- Debyser Zeger (2003). "A Short Course on Virology / Vectorology / Gene Therapy" (PDF). Current Gene Therapy. 3 (6): 495–99. doi:10.2174/1566523034578122. PMID 14683447. Archived from the original (PDF) on 2008-08-02.
- Kolata, Gina (2005-10-06). "Experts Unlock Clues to Spread of 1918 Flu Virus". The New York Times. ISSN 0362-4331. Retrieved 2008-02-03.
- Stem Cells – This Time without the Cancer, Scientific American News, 30 November 2007
- "Biggest Known Virus Yields First-Ever Virophage". Microbe Magazine. November 2008. Archived from the original on July 22, 2011.
- "Virus hiding in our genome protects early human embryos". New Scientist. 20 April 2015.
- H. Tang et al., Human Organs-on-Chips for Virology, Trends in Microbiology (2020)
Further reading
- Baron, Samuel, ed. (1996). "Section 2: Virology". Medical Microbiology (4th ed.). (freely searchable online book)
- Coffin; Hughes; Varmus (1997). Retroviruses. (freely searchable online book)
- Villarreal, L. P. (2005). Viruses and the Evolution of Life. Washington DC: ASM Press. ISBN 1-55581-309-7.
External links
- ICTV: International Committee on Taxonomy of Viruses – searchable virus taxonomy, updated versions in 2007 and 2008. Explanations of the virus species concept and viral taxonomy.
- David Sander: All the Virology on the WWW – collection of links, pictures, lecture notes. Many of the links on this site are broken and it does not appear to be being maintained.
- Online lectures in virology University of South Carolina
- Microbes.info is a microbiology information portal containing a vast collection of resources including articles, news, frequently asked questions, and links pertaining to the field of microbiology.
- MicrobiologyBytes: Origins of Virology
- MicrobiologyBytes: The Virology Time Machine
- Timeline of the history of virology, from the Washington University in St. Louis.
- Wong's Virology.
- Vaccine Research Center (VRC) – Information concerning vaccine research studies
- This Week in Virology Podcast by Vincent Racaniello
- Virulogy, Ton E. van den Bogaard University Maastricht, the Netherlands,