Succinate dehydrogenase
Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory Complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain.[1] Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.[2]
| succinate dehydrogenase (succinate-ubiquinone oxidoreductase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 The structure of SQR in a phospholipid membrane. SdhA, SdhB, SdhC and SdhD | |||||||||
| Identifiers | |||||||||
| EC number | 1.3.5.1 | ||||||||
| CAS number | 9028-11-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Succinate dehydrogenase | |
|---|---|
| Identifiers | |
| Symbol | Respiratory complex II |
| OPM superfamily | 3 |
| OPM protein | 1zoy |
| Membranome | 656 |
In step 6 of the citric acid cycle, SQR catalyzes the oxidation of succinate to fumarate with the reduction of ubiquinone to ubiquinol. This occurs in the inner mitochondrial membrane by coupling the two reactions together.
Structure
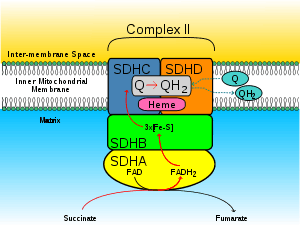
Subunits
Mitochondrial and many bacterial SQRs are composed of four structurally different subunits: two hydrophilic and two hydrophobic. The first two subunits, a flavoprotein (SdhA) and an iron-sulfur protein (SdhB), form a hydrophilic head where enzymatic activity of the complex takes place. SdhA contains a covalently attached flavin adenine dinucleotide (FAD) cofactor and the succinate binding site and SdhB contains three iron-sulfur clusters: [2Fe-2S], [4Fe-4S], and [3Fe-4S]. The second two subunits are hydrophobic membrane anchor subunits, SdhC and SdhD. Human mitochondria contain two distinct isoforms of SdhA (Fp subunits type I and type II), these isoforms are also found in Ascaris suum and Caenorhabditis elegans.[3] The subunits form a membrane-bound cytochrome b complex with six transmembrane helices containing one heme b group and a ubiquinone-binding site. Two phospholipid molecules, one cardiolipin and one phosphatidylethanolamine, are also found in the SdhC and SdhD subunits (not shown in the image). They serve to occupy the hydrophobic space below the heme b. These subunits are displayed in the attached image. SdhA is green, SdhB is teal, SdhC is fuchsia, and SdhD is yellow. Around SdhC and SdhD is a phospholipid membrane with the intermembrane space at the top of the image.[4]
Table of subunit composition[5]
| No. | Subunit name | Human protein | Protein description from UniProt | Pfam family with Human protein |
|---|---|---|---|---|
| 1 | SdhA | SDHA_HUMAN | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Pfam PF00890, Pfam PF02910 |
| 2 | SdhB | SDHB_HUMAN | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | Pfam PF13085, Pfam PF13183 |
| 3 | SdhC | C560_HUMAN | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial | Pfam PF01127 |
| 4 | SdhD | DHSD_HUMAN | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit, mitochondrial | Pfam PF05328 |
Ubiquinone binding site
Two distinctive ubiquinone binding sites can be recognized on mammalian SDH – matrix-proximal QP and matrix-distal QD. Ubiquinone binding site Qp, which shows higher affinity to ubiquinone, is located in a gap composed of SdhB, SdhC, and SdhD. Ubiquinone is stabilized by the side chains of His207 of subunit B, Ser27 and Arg31 of subunit C, and Tyr83 of subunit D. The quinone ring is surrounded by Ile28 of subunit C and Pro160 of subunit B. These residues, along with Il209, Trp163, and Trp164 of subunit B, and Ser27 (C atom) of subunit C, form the hydrophobic environment of the quinone-binding pocket Qp.[6] In contrast, ubiquinone binding site QD, which lies closer to inter-membrane space, is composed of SdhD only and has lower affinity to ubiquinone.[7]
Succinate binding site
SdhA provides the binding site for the oxidation of succinate. The side chains Thr254, His354, and Arg399 of subunit A stabilize the molecule while FAD oxidizes and carries the electrons to the first of the iron-sulfur clusters, [2Fe-2S].[8] This can be seen in image 5.
Redox centers
The succinate-binding site and ubiquinone-binding site are connected by a chain of redox centers including FAD and the iron-sulfur clusters. This chain extends over 40 Å through the enzyme monomer. All edge-to-edge distances between the centers are less than the suggested 14 Å limit for physiological electron transfer.[4] This electron transfer is demonstrated in image 8.
Subunit E
| SdhE | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Solution NMR structure of protein NMA1147 from Neisseria meningitidis. Northeast structural genomics consortium target mr19 | |||||||||
| Identifiers | |||||||||
| Symbol | SdhE | ||||||||
| Pfam | PF03937 | ||||||||
| InterPro | IPR005631 | ||||||||
| |||||||||
In molecular biology, the protein domain named Sdh5 is also named SdhE which stands for succinate dehydrogenase protein E. In the past, it has also been named YgfY and DUF339.[9] Another name for sdhE is succinate dehydrogenase assembly factor 2 (sdhaf2).[10] This protein belongs to a group of highly conserved small proteins found in both eukaryotes and prokaryotes, including NMA1147 from Neisseria meningitidis [11] and YgfY from Escherichia coli.[12] The SdhE protein is found on the mitochondrial membrane is it is important for creating energy via a process named the electron transport chain.[9]
Function
The function of SDHE has been described as a flavinator of succinate dehydrogenase. SdhE works as a co-factor chaperone that incorporates FAD into SdhA. This results in SdhA flavinylation which is required for the proper function succinate dehydrogenase. More recent scientific studies indicate that SdhE is required by bacteria in order to grow on succinate, using succinate as its only source of carbon and additionally for the function, of succinate dehydrogenase, a vital component of the electron transport chain which produces energy.[9]
Structure
The structure of these proteins consists of a complex bundle of five alpha-helices, which is composed of an up-down 3-helix bundle plus an orthogonal 2-helix bundle.[12]
Protein interactions
SdhE interacts with the catalytic subunit of the succinate dehydrogenase (SDH) complex.[13]
Human disease
The human gene named SDH5, encodes for the SdhE protein. The gene itself is located in the chromosomal position 11q13.1. Loss-of-function mutations result in paraganglioma, a neuroendocrine tumour.[13]
History
The recent studies which suggest SdhE is required for bacterial flavinylation contradict previous thoughts on SdhE. It was originally proposed that FAD incorporation into bacterial flavoproteins was an autocatalytic process. Recent studies now argue that SdhE is the first protein to be identified as required for flavinylation in bacteria. Historically, the SdhE protein was once considered a hypothetical protein.[9] YgfY was also thought to be involved in transcriptional regulation.[12]
Assembly and maturation
All subunits of human mitochondrial SDH are encoded in nuclear genome. After translation, SDHA subunit is translocated as apoprotein into the mitochondrial matrix. Subsequently, one of the first steps is covalent binding of the FAD cofactor (flavinylation). This process seems to be regulated by some of the tricarboxylic acid cycle intermediates. Specifically, succinate, isocitrate and citrate stimulate flavinylation of the SDHA.[14] In case of eukaryotic Sdh1 (SDHA in mammals), another protein is required for process of FAD incorporation – namely Sdh5 in yeast, succinate dehydrogenase assembly factor 2 (SDHAF2) in mammal cells.
Before forming a heterodimer with subunit SDHB, some portion of SDHA with covalently bound FAD appears to interact with other assembly factor – SDHAF4 (Sdh8 in yeast). Unbound flavinylated SDHA dimerizes with SDHAF4 which serves as a chaperone. Studies suggest that formation of SDHA-SDHB dimer is impaired in absence of SDHAF4 so the chaperon-like assembly factor might facilitate interaction of the subunits. Moreover, SDHAF4 seems to prevent ROS generation via accepting electrons from succinate which can be still oxidized by unbound monomeric SDHA subunit.[7]
Fe-S prosthetic groups of the subunit SDHB are being preformed in the mitochondrial matrix by protein complex ISU. The complex is also thought to be capable of inserting the iron-sulphur clusters in SDHB during its maturation. The studies suggest that Fe-S cluster insertion precedes SDHA-SDHB dimer forming. Such incorporation requires reduction of cysteine residues within active site of SDHB. Both reduced cysteine residues and already incorporated Fe-S clusters are highly susceptible to ROS damage. Two more SDH assembly factors, SDHAF1 (Sdh6) and SDHAF3 (Sdh7 in yeast), seem to be involved in SDHB maturation in way of protecting the subunit or dimer SDHA-SDHB from Fe-S cluster damage caused by ROS.[7]
Assembly of the hydrophobic anchor consisting of subunits SDHC and SDHD remains unclear. Especially in case of heme b insertion and even its function. Heme b prosthetic group does not appear to be part of electron transporting pathway within the complex II.[15] The cofactor rather maintains the anchor stability.
Mechanism
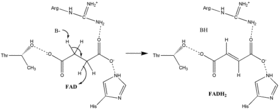
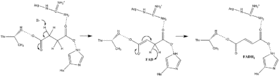
Succinate oxidation
Little is known about the exact succinate oxidation mechanism. However, the crystal structure shows that FAD, Glu255, Arg286, and His242 of subunit A (not shown) are good candidates for the initial deprotonation step. Thereafter, there are two possible elimination mechanisms: E2 or E1cb. In the E2 elimination, the mechanism is concerted. The basic residue or cofactor deprotonates the alpha carbon, and FAD accepts the hydride from the beta carbon, oxidizing the bound succinate to fumarate—refer to image 6. In E1cb, an enolate intermediate is formed, shown in image 7, before FAD accepts the hydride. Further research is required to determine which elimination mechanism succinate undergoes in Succinate Dehydrogenase. Oxidized fumarate, now loosely bound to the active site, is free to exit the protein.
Electron tunneling
After the electrons are derived from succinate oxidation via FAD, they tunnel along the [Fe-S] relay until they reach the [3Fe-4S] cluster. These electrons are subsequently transferred to an awaiting ubiquinone molecule within the active site. The Iron-Sulfur electron tunneling system is shown in image 9.
Ubiquinone reduction
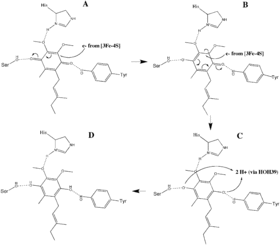
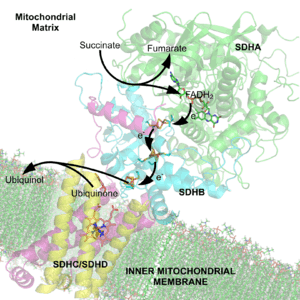
The O1 carbonyl oxygen of ubiquinone is oriented at the active site (image 4) by hydrogen bond interactions with Tyr83 of subunit D. The presence of electrons in the [3Fe-4S] iron sulphur cluster induces the movement of ubiquinone into a second orientation. This facilitates a second hydrogen bond interaction between the O4 carbonyl group of ubiquinone and Ser27 of subunit C. Following the first single electron reduction step, a semiquinone radical species is formed. The second electron arrives from the [3Fe-4S] cluster to provide full reduction of the ubiquinone to ubiquinol. This mechanism of the ubiquinone reduction is shown in image 8.
Heme prosthetic group
Although the functionality of the heme in succinate dehydrogenase is still being researched, some studies have asserted that the first electron delivered to ubiquinone via [3Fe-4S] may tunnel back and forth between the heme and the ubiquinone intermediate. In this way, the heme cofactor acts as an electron sink. Its role is to prevent the interaction of the intermediate with molecular oxygen to produce reactive oxygen species (ROS). The heme group, relative to ubiquinone, is shown in image 4.
It has also been proposed that a gating mechanism may be in place to prevent the electrons from tunneling directly to the heme from the [3Fe-4S] cluster. A potential candidate is residue His207, which lies directly between the cluster and the heme. His207 of subunit B is in direct proximity to the [3Fe-4S] cluster, the bound ubiquinone, and the heme; and could modulate electron flow between these redox centers.[16]
Proton transfer
To fully reduce the quinone in SQR, two electrons as well as two protons are needed. It has been argued that a water molecule (HOH39) arrives at the active site and is coordinated by His207 of subunit B, Arg31 of subunit C, and Asp82 of subunit D. The semiquinone species is protonated by protons delivered from HOH39, completing the ubiquinone reduction to ubiquinol. His207 and Asp82 most likely facilitate this process. Other studies claim that Tyr83 of subunit D is coordinated to a nearby histidine as well as the O1 carbonyl oxygen of ubiquinone. The histidine residue decreases the pKa of tyrosine, making it more suitable to donate its proton to the reduced ubiquinone intermediate.
Inhibitors
There are two distinct classes of inhibitors of complex II: those that bind in the succinate pocket and those that bind in the ubiquinone pocket. Ubiquinone type inhibitors include carboxin and thenoyltrifluoroacetone. Succinate-analogue inhibitors include the synthetic compound malonate as well as the TCA cycle intermediates, malate and oxaloacetate. Indeed, oxaloacetate is one of the most potent inhibitors of Complex II. Why a common TCA cycle intermediate would inhibit Complex II is not entirely understood, though it may exert a protective role in minimizing reverse-electron transfer mediated production of superoxide by Complex I.[17] Atpenin 5a are highly potent Complex II inhibitors mimicking ubiquinone binding.
Ubiquinone type inhibitors have been used as fungicides in agriculture since the 1960s. Carboxin was mainly used to control disease caused by basidiomycetes such as stem rusts and Rhizoctonia diseases. More recently, other compounds with a broader spectrum against a range of plant pathogens have been developed including boscalid, penthiopyrad and fluopyram.[18] Some agriculturally important fungi are not sensitive towards members of the new generation of ubiquinone type inhibitors [19]
Role in disease
The fundamental role of succinate-coenzyme Q reductase in the electron transfer chain of mitochondria makes it vital in most multicellular organisms, removal of this enzyme from the genome has also been shown to be lethal at the embryonic stage in mice.
- SdhA mutations can lead to Leigh syndrome, mitochondrial encephalopathy, and optic atrophy.
- SdhB mutations can lead to tumorogenesis in chromaffin cells, causing a class of tumors known as succinate dehydrogenase deficient including hereditary paraganglioma and hereditary pheochromocytoma, succinate dehydrogenase deficient renal carcinoma and succinate dehydrogenase deficient gastrointestinal stromal tumor (GIST).[20] Tumors tend to be malignant. It can also lead to decreased life-span and increased production of superoxide ions.
- SdhC mutations can lead to decreased life-span, increased production of superoxide ions, hereditary paraganglioma and hereditary pheochromocytoma. Tumors tend to be benign. These mutations are uncommon.
- SdhD mutations can lead to hereditary paraganglioma and hereditary pheochromocytoma. Tumors tend to be benign, and occur often in the head and neck regions. These mutations can also decrease life-span and increase production of superoxide ions.
Mammalian succinate dehydrogenase functions not only in mitochondrial energy generation, but also has a role in oxygen sensing and tumor suppression; and, therefore, is the object of ongoing research.
Reduced levels of the mitochondrial enzyme succinate dehydrogenase (SDH), the main element of complex II, are observed post mortem in the brains of patients with Huntington's Disease, and energy metabolism defects have been identified in both presymptomatic and symptomatic HD patients.[21]
See also
References
- Oyedotun KS, Lemire BD (March 2004). "The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies". The Journal of Biological Chemistry. 279 (10): 9424–31. doi:10.1074/jbc.M311876200. PMID 14672929.
- webmaster (2009-03-04). "Using Histochemistry to Determine Muscle Properties". Succinate Dehydrogenase: Identifying Oxidative Potential. University of California, San Diego. Retrieved 2017-12-27.
- Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K (August 2003). "Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase)". Journal of Biochemistry. 134 (2): 191–5. doi:10.1093/jb/mvg144. PMID 12966066.
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, et al. (January 2003). "Architecture of succinate dehydrogenase and reactive oxygen species generation". Science. 299 (5607): 700–4. Bibcode:2003Sci...299..700Y. doi:10.1126/science.1079605. PMID 12560550.
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, et al. (July 2005). "Crystal structure of mitochondrial respiratory membrane protein complex II". Cell. 121 (7): 1043–57. doi:10.1016/j.cell.2005.05.025. PMID 15989954.
- Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, et al. (March 2006). "Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction". The Journal of Biological Chemistry. 281 (11): 7309–16. doi:10.1074/jbc.M508173200. PMID 16407191.
- Van Vranken JG, Na U, Winge DR, Rutter J (December 2014). "Protein-mediated assembly of succinate dehydrogenase and its cofactors". Critical Reviews in Biochemistry and Molecular Biology. 50 (2): 168–80. doi:10.3109/10409238.2014.990556. PMC 4653115. PMID 25488574.
- Kenney WC (April 1975). "The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase". The Journal of Biological Chemistry. 250 (8): 3089–94. PMID 235539.
- McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC (2012). "SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria". J Biol Chem. 287 (22): 18418–28. doi:10.1074/jbc.M111.293803. PMC 3365757. PMID 22474332.
- https://www.genecards.org/cgi-bin/carddisp.pl?gene=SDHAF2
- Liu G, Sukumaran DK, Xu D, Chiang Y, Acton T, Goldsmith-Fischman S, Honig B, Montelione GT, Szyperski T (May 2004). "NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle". Proteins. 55 (3): 756–8. doi:10.1002/prot.20009. PMID 15103637.
- Lim K, Doseeva V, Demirkan ES, Pullalarevu S, Krajewski W, Galkin A, Howard A, Herzberg O (February 2005). "Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation". Proteins. 58 (3): 759–63. doi:10.1002/prot.20337. PMID 15593094.
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, et al. (2009). "SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma". Science. 325 (5944): 1139–42. Bibcode:2009Sci...325.1139H. doi:10.1126/science.1175689. PMC 3881419. PMID 19628817.
- Brandsch R, Bichler V (June 1989). "Covalent cofactor binding to flavoenzymes requires specific effectors". European Journal of Biochemistry. 182 (1): 125–8. doi:10.1111/j.1432-1033.1989.tb14808.x. PMID 2659351.
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, et al. (July 2005). "Crystal structure of mitochondrial respiratory membrane protein complex II". Cell. 121 (7): 1043–57. doi:10.1016/j.cell.2005.05.025. PMID 15989954.
- Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH (October 2006). "The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b". The Journal of Biological Chemistry. 281 (43): 32310–7. doi:10.1074/jbc.M607476200. PMID 16950775.
- Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, Van Remmen H (January 2008). "High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates". The Biochemical Journal. 409 (2): 491–9. doi:10.1042/BJ20071162. PMID 17916065.
- Avenot, H. F.; Michailides, T. J. (2010). "Progress in understanding molecular mechanisms and evolution of resistance to succinate dehydrogenase inhibiting (SDHI) fungicides in phytopathogenic fungi". Crop Protection. 29 (7): 643. doi:10.1016/j.cropro.2010.02.019.
- Dubos T, Pasquali M, Pogoda F, Casanova A, Hoffmann L, Beyer M (January 2013). "Differences between the succinate dehydrogenase sequences of isopyrazam sensitive Zymoseptoria tritici and insensitive Fusarium graminearum strains". Pesticide Biochemistry and Physiology. 105 (1): 28–35. doi:10.1016/j.pestbp.2012.11.004. PMID 24238287.
- Barletta JA, Hornick JL (July 2012). "Succinate dehydrogenase-deficient tumors: diagnostic advances and clinical implications". Advances in Anatomic Pathology. 19 (4): 193–203. doi:10.1097/PAP.0b013e31825c6bc6. PMID 22692282.
- Skillings EA, Morton AJ (2016). "Delayed Onset and Reduced Cognitive Deficits through Pre-Conditioning with 3-Nitropropionic Acid is Dependent on Sex and CAG Repeat Length in the R6/2 Mouse Model of Huntington's Disease". Journal of Huntington's Disease. 5 (1): 19–32. doi:10.3233/JHD-160189. PMID 27031731.