Meta-analysis
A meta-analysis is a statistical analysis that combines the results of multiple scientific studies. Meta-analysis can be performed when there are multiple scientific studies addressing the same question, with each individual study reporting measurements that are expected to have some degree of error. The aim then is to use approaches from statistics to derive a pooled estimate closest to the unknown common truth based on how this error is perceived.
Existing methods for meta-analysis yield a weighted average from the results of the individual studies, and what differs is the manner in which these weights are allocated and also the manner in which the uncertainty is computed around the point estimate thus generated. In addition to providing an estimate of the unknown common truth, meta-analysis has the capacity to contrast results from different studies and identify patterns among study results, sources of disagreement among those results, or other interesting relationships that may come to light in the context of multiple studies.[1]
A key benefit of this approach is the aggregation of information leading to a higher statistical power and more robust point estimate than is possible from the measure derived from any individual study. However, in performing a meta-analysis, an investigator must make choices which can affect the results, including deciding how to search for studies, selecting studies based on a set of objective criteria, dealing with incomplete data, analyzing the data, and accounting for or choosing not to account for publication bias.[2] Judgment calls made in completing a meta-analysis may affect the results. For example, Wanous and colleagues examined four pairs of meta-analyses on the four topics of (a) job performance and satisfaction relationship, (b) realistic job previews, (c) correlates of role conflict and ambiguity, and (d) the job satisfaction and absenteeism relationship, and illustrated how various judgement calls made by the researchers produced different results.[3]
Meta-analyses are often, but not always, important components of a systematic review procedure. For instance, a meta-analysis may be conducted on several clinical trials of a medical treatment, in an effort to obtain a better understanding of how well the treatment works. Here it is convenient to follow the terminology used by the Cochrane Collaboration,[4] and use "meta-analysis" to refer to statistical methods of combining evidence, leaving other aspects of 'research synthesis' or 'evidence synthesis', such as combining information from qualitative studies, for the more general context of systematic reviews. A meta-analysis is a secondary source.[5][6]
History
The historical roots of meta-analysis can be traced back to 17th century studies of astronomy,[7] while a paper published in 1904 by the statistician Karl Pearson in the British Medical Journal[8] which collated data from several studies of typhoid inoculation is seen as the first time a meta-analytic approach was used to aggregate the outcomes of multiple clinical studies.[9][10] The first meta-analysis of all conceptually identical experiments concerning a particular research issue, and conducted by independent researchers, has been identified as the 1940 book-length publication Extrasensory Perception After Sixty Years, authored by Duke University psychologists J. G. Pratt, J. B. Rhine, and associates.[11] This encompassed a review of 145 reports on ESP experiments published from 1882 to 1939, and included an estimate of the influence of unpublished papers on the overall effect (the file-drawer problem). Although meta-analysis is widely used in epidemiology and evidence-based medicine today, a meta-analysis of a medical treatment was not published until 1955. In the 1970s, more sophisticated analytical techniques were introduced in educational research, starting with the work of Gene V. Glass, Frank L. Schmidt and John E. Hunter.
The term "meta-analysis" was coined in 1976 by the statistician Gene V. Glass,[12] who stated "my major interest currently is in what we have come to call ...the meta-analysis of research. The term is a bit grand, but it is precise and apt ... Meta-analysis refers to the analysis of analyses". Although this led to him being widely recognized as the modern founder of the method, the methodology behind what he termed "meta-analysis" predates his work by several decades.[13][14] The statistical theory surrounding meta-analysis was greatly advanced by the work of Nambury S. Raju, Larry V. Hedges, Harris Cooper, Ingram Olkin, John E. Hunter, Jacob Cohen, Thomas C. Chalmers, Robert Rosenthal, Frank L. Schmidt, and Douglas G. Bonett.
Steps in a meta-analysis
A meta-analysis is usually preceded by a systematic review, as this allows identification and critical appraisal of all the relevant evidence (thereby limiting the risk of bias in summary estimates). The general steps are then as follows:
- Formulation of the research question, e.g. using the PICO model (Population, Intervention, Comparison, Outcome).
- Search of literature
- Selection of studies ('incorporation criteria')
- Based on quality criteria, e.g. the requirement of randomization and blinding in a clinical trial
- Selection of specific studies on a well-specified subject, e.g. the treatment of breast cancer.
- Decide whether unpublished studies are included to avoid publication bias (file drawer problem)
- Decide which dependent variables or summary measures are allowed. For instance, when considering a meta-analysis of published (aggregate) data:
- Differences (discrete data)
- Means (continuous data)
- Hedges' g is a popular summary measure for continuous data that is standardized in order to eliminate scale differences, but it incorporates an index of variation between groups:
- in which is the treatment mean, is the control mean, the pooled variance.
- Selection of a meta-analysis model, e.g. fixed effect or random effects meta-analysis.
- Examine sources of between-study heterogeneity, e.g. using subgroup analysis or meta-regression.
Formal guidance for the conduct and reporting of meta-analyses is provided by the Cochrane Handbook.
For reporting guidelines, see the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.[15]
Methods and assumptions
Approaches
In general, two types of evidence can be distinguished when performing a meta-analysis: individual participant data (IPD), and aggregate data (AD). The aggregate data can be direct or indirect.
AD is more commonly available (e.g. from the literature) and typically represents summary estimates such as odds ratios or relative risks. This can be directly synthesized across conceptually similar studies using several approaches (see below). On the other hand, indirect aggregate data measures the effect of two treatments that were each compared against a similar control group in a meta-analysis. For example, if treatment A and treatment B were directly compared vs placebo in separate meta-analyses, we can use these two pooled results to get an estimate of the effects of A vs B in an indirect comparison as effect A vs Placebo minus effect B vs Placebo.
IPD evidence represents raw data as collected by the study centers. This distinction has raised the need for different meta-analytic methods when evidence synthesis is desired, and has led to the development of one-stage and two-stage methods.[16] In one-stage methods the IPD from all studies are modeled simultaneously whilst accounting for the clustering of participants within studies. Two-stage methods first compute summary statistics for AD from each study and then calculate overall statistics as a weighted average of the study statistics. By reducing IPD to AD, two-stage methods can also be applied when IPD is available; this makes them an appealing choice when performing a meta-analysis. Although it is conventionally believed that one-stage and two-stage methods yield similar results, recent studies have shown that they may occasionally lead to different conclusions.[17][18]
Statistical models for aggregate data
Direct evidence: Models incorporating study effects only
Fixed effects model
The fixed effect model provides a weighted average of a series of study estimates. The inverse of the estimates' variance is commonly used as study weight, so that larger studies tend to contribute more than smaller studies to the weighted average. Consequently, when studies within a meta-analysis are dominated by a very large study, the findings from smaller studies are practically ignored.[19] Most importantly, the fixed effects model assumes that all included studies investigate the same population, use the same variable and outcome definitions, etc. This assumption is typically unrealistic as research is often prone to several sources of heterogeneity; e.g. treatment effects may differ according to locale, dosage levels, study conditions, ...
Random effects model
A common model used to synthesize heterogeneous research is the random effects model of meta-analysis. This is simply the weighted average of the effect sizes of a group of studies. The weight that is applied in this process of weighted averaging with a random effects meta-analysis is achieved in two steps:[20]
- Step 1: Inverse variance weighting
- Step 2: Un-weighting of this inverse variance weighting by applying a random effects variance component (REVC) that is simply derived from the extent of variability of the effect sizes of the underlying studies.
This means that the greater this variability in effect sizes (otherwise known as heterogeneity), the greater the un-weighting and this can reach a point when the random effects meta-analysis result becomes simply the un-weighted average effect size across the studies. At the other extreme, when all effect sizes are similar (or variability does not exceed sampling error), no REVC is applied and the random effects meta-analysis defaults to simply a fixed effect meta-analysis (only inverse variance weighting).
The extent of this reversal is solely dependent on two factors:[21]
- Heterogeneity of precision
- Heterogeneity of effect size
Since neither of these factors automatically indicates a faulty larger study or more reliable smaller studies, the re-distribution of weights under this model will not bear a relationship to what these studies actually might offer. Indeed, it has been demonstrated that redistribution of weights is simply in one direction from larger to smaller studies as heterogeneity increases until eventually all studies have equal weight and no more redistribution is possible.[21] Another issue with the random effects model is that the most commonly used confidence intervals generally do not retain their coverage probability above the specified nominal level and thus substantially underestimate the statistical error and are potentially overconfident in their conclusions.[22][23] Several fixes have been suggested[24][25] but the debate continues on.[23][26] A further concern is that the average treatment effect can sometimes be even less conservative compared to the fixed effect model[27] and therefore misleading in practice. One interpretational fix that has been suggested is to create a prediction interval around the random effects estimate to portray the range of possible effects in practice.[28] However, an assumption behind the calculation of such a prediction interval is that trials are considered more or less homogeneous entities and that included patient populations and comparator treatments should be considered exchangeable[29] and this is usually unattainable in practice.
The most widely used method to estimate between studies variance (REVC) is the DerSimonian-Laird (DL) approach.[30] Several advanced iterative (and computationally expensive) techniques for computing the between studies variance exist (such as maximum likelihood, profile likelihood and restricted maximum likelihood methods) and random effects models using these methods can be run in Stata with the metaan command.[31] The metaan command must be distinguished from the classic metan (single "a") command in Stata that uses the DL estimator. These advanced methods have also been implemented in a free and easy to use Microsoft Excel add-on, MetaEasy.[32][33] However, a comparison between these advanced methods and the DL method of computing the between studies variance demonstrated that there is little to gain and DL is quite adequate in most scenarios.[34][35]
However, most meta-analyses include between 2 and 4 studies and such a sample is more often than not inadequate to accurately estimate heterogeneity. Thus it appears that in small meta-analyses, an incorrect zero between study variance estimate is obtained, leading to a false homogeneity assumption. Overall, it appears that heterogeneity is being consistently underestimated in meta-analyses and sensitivity analyses in which high heterogeneity levels are assumed could be informative.[36] These random effects models and software packages mentioned above relate to study-aggregate meta-analyses and researchers wishing to conduct individual patient data (IPD) meta-analyses need to consider mixed-effects modelling approaches.[37]
IVhet model
Doi & Barendregt working in collaboration with Khan, Thalib and Williams (from the University of Queensland, University of Southern Queensland and Kuwait University), have created an inverse variance quasi likelihood based alternative (IVhet) to the random effects (RE) model for which details are available online.[38] This was incorporated into MetaXL version 2.0,[39] a free Microsoft excel add-in for meta-analysis produced by Epigear International Pty Ltd, and made available on 5 April 2014. The authors state that a clear advantage of this model is that it resolves the two main problems of the random effects model. The first advantage of the IVhet model is that coverage remains at the nominal (usually 95%) level for the confidence interval unlike the random effects model which drops in coverage with increasing heterogeneity.[22][23] The second advantage is that the IVhet model maintains the inverse variance weights of individual studies, unlike the RE model which gives small studies more weight (and therefore larger studies less) with increasing heterogeneity. When heterogeneity becomes large, the individual study weights under the RE model become equal and thus the RE model returns an arithmetic mean rather than a weighted average. This side-effect of the RE model does not occur with the IVhet model which thus differs from the RE model estimate in two perspectives:[38] Pooled estimates will favor larger trials (as opposed to penalizing larger trials in the RE model) and will have a confidence interval that remains within the nominal coverage under uncertainty (heterogeneity). Doi & Barendregt suggest that while the RE model provides an alternative method of pooling the study data, their simulation results[40] demonstrate that using a more specified probability model with untenable assumptions, as with the RE model, does not necessarily provide better results. The latter study also reports that the IVhet model resolves the problems related to underestimation of the statistical error, poor coverage of the confidence interval and increased MSE seen with the random effects model and the authors conclude that researchers should henceforth abandon use of the random effects model in meta-analysis. While their data is compelling, the ramifications (in terms of the magnitude of spuriously positive results within the Cochrane database) are huge and thus accepting this conclusion requires careful independent confirmation. The availability of a free software (MetaXL)[39] that runs the IVhet model (and all other models for comparison) facilitates this for the research community.
Direct evidence: Models incorporating additional information
Quality effects model
Doi and Thalib originally introduced the quality effects model.[41] They[42] introduced a new approach to adjustment for inter-study variability by incorporating the contribution of variance due to a relevant component (quality) in addition to the contribution of variance due to random error that is used in any fixed effects meta-analysis model to generate weights for each study. The strength of the quality effects meta-analysis is that it allows available methodological evidence to be used over subjective random effects, and thereby helps to close the damaging gap which has opened up between methodology and statistics in clinical research. To do this a synthetic bias variance is computed based on quality information to adjust inverse variance weights and the quality adjusted weight of the ith study is introduced.[41] These adjusted weights are then used in meta-analysis. In other words, if study i is of good quality and other studies are of poor quality, a proportion of their quality adjusted weights is mathematically redistributed to study i giving it more weight towards the overall effect size. As studies become increasingly similar in terms of quality, re-distribution becomes progressively less and ceases when all studies are of equal quality (in the case of equal quality, the quality effects model defaults to the IVhet model – see previous section). A recent evaluation of the quality effects model (with some updates) demonstrates that despite the subjectivity of quality assessment, the performance (MSE and true variance under simulation) is superior to that achievable with the random effects model.[43][44] This model thus replaces the untenable interpretations that abound in the literature and a software is available to explore this method further.[39]
Indirect evidence: Network meta-analysis methods
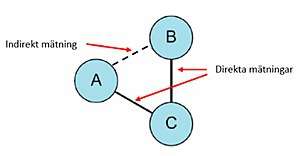
Indirect comparison meta-analysis methods (also called network meta-analyses, in particular when multiple treatments are assessed simultaneously) generally use two main methodologies. First, is the Bucher method[45] which is a single or repeated comparison of a closed loop of three-treatments such that one of them is common to the two studies and forms the node where the loop begins and ends. Therefore, multiple two-by-two comparisons (3-treatment loops) are needed to compare multiple treatments. This methodology requires that trials with more than two arms have two arms only selected as independent pair-wise comparisons are required. The alternative methodology uses complex statistical modelling to include the multiple arm trials and comparisons simultaneously between all competing treatments. These have been executed using Bayesian methods, mixed linear models and meta-regression approaches.
Bayesian framework
Specifying a Bayesian network meta-analysis model involves writing a directed acyclic graph (DAG) model for general-purpose Markov chain Monte Carlo (MCMC) software such as WinBUGS.[46] In addition, prior distributions have to be specified for a number of the parameters, and the data have to be supplied in a specific format.[46] Together, the DAG, priors, and data form a Bayesian hierarchical model. To complicate matters further, because of the nature of MCMC estimation, overdispersed starting values have to be chosen for a number of independent chains so that convergence can be assessed.[47] Currently, there is no software that automatically generates such models, although there are some tools to aid in the process. The complexity of the Bayesian approach has limited usage of this methodology. Methodology for automation of this method has been suggested[48] but requires that arm-level outcome data are available, and this is usually unavailable. Great claims are sometimes made for the inherent ability of the Bayesian framework to handle network meta-analysis and its greater flexibility. However, this choice of implementation of framework for inference, Bayesian or frequentist, may be less important than other choices regarding the modeling of effects[49] (see discussion on models above).
Frequentist multivariate framework
On the other hand, the frequentist multivariate methods involve approximations and assumptions that are not stated explicitly or verified when the methods are applied (see discussion on meta-analysis models above). For example, the mvmeta package for Stata enables network meta-analysis in a frequentist framework.[50] However, if there is no common comparator in the network, then this has to be handled by augmenting the dataset with fictional arms with high variance, which is not very objective and requires a decision as to what constitutes a sufficiently high variance.[51] The other issue is use of the random effects model in both this frequentist framework and the Bayesian framework. Senn advises analysts to be cautious about interpreting the 'random effects' analysis since only one random effect is allowed for but one could envisage many.[49] Senn goes on to say that it is rather naıve, even in the case where only two treatments are being compared to assume that random-effects analysis accounts for all uncertainty about the way effects can vary from trial to trial. Newer models of meta-analysis such as those discussed above would certainly help alleviate this situation and have been implemented in the next framework.
Generalized pairwise modelling framework
An approach that has been tried since the late 1990s is the implementation of the multiple three-treatment closed-loop analysis. This has not been popular because the process rapidly becomes overwhelming as network complexity increases. Development in this area was then abandoned in favor of the Bayesian and multivariate frequentist methods which emerged as alternatives. Very recently, automation of the three-treatment closed loop method has been developed for complex networks by some researchers[38] as a way to make this methodology available to the mainstream research community. This proposal does restrict each trial to two interventions, but also introduces a workaround for multiple arm trials: a different fixed control node can be selected in different runs. It also utilizes robust meta-analysis methods so that many of the problems highlighted above are avoided. Further research around this framework is required to determine if this is indeed superior to the Bayesian or multivariate frequentist frameworks. Researchers willing to try this out have access to this framework through a free software.[39]
Tailored meta-analysis
Another form of additional information comes from the intended setting. If the target setting for applying the meta-analysis results is known then it may be possible to use data from the setting to tailor the results thus producing a 'tailored meta-analysis'.,[52][53] This has been used in test accuracy meta-analyses, where empirical knowledge of the test positive rate and the prevalence have been used to derive a region in Receiver Operating Characteristic (ROC) space known as an 'applicable region'. Studies are then selected for the target setting based on comparison with this region and aggregated to produce a summary estimate which is tailored to the target setting.
Aggregating IPD and AD
Meta-analysis can also be applied to combine IPD and AD. This is convenient when the researchers who conduct the analysis have their own raw data while collecting aggregate or summary data from the literature. The generalized integration model (GIM)[54] is a generalization of the meta-analysis. It allows that the model fitted on the individual participant data (IPD) is different from the ones used to compute the aggregate data (AD). GIM can be viewed as a model calibration method for integrating information with more flexibility.
Validation of meta-analysis results
The meta-analysis estimate represents a weighted average across studies and when there is heterogeneity this may result in the summary estimate not being representative of individual studies. Qualitative appraisal of the primary studies using established tools can uncover potential biases,[55][56] but does not quantify the aggregate effect of these biases on the summary estimate. Although the meta-analysis result could be compared with an independent prospective primary study, such external validation is often impractical. This has led to the development of methods that exploit a form of leave-one-out cross validation, sometimes referred to as internal-external cross validation (IOCV).[57] Here each of the k included studies in turn is omitted and compared with the summary estimate derived from aggregating the remaining k- 1 studies. A general validation statistic, Vn based on IOCV has been developed to measure the statistical validity of meta-analysis results.[58] For test accuracy and prediction, particularly when there are multivariate effects, other approaches which seek to estimate the prediction error have also been proposed.[59]
Challenges
A meta-analysis of several small studies does not always predict the results of a single large study.[60] Some have argued that a weakness of the method is that sources of bias are not controlled by the method: a good meta-analysis cannot correct for poor design or bias in the original studies.[61] This would mean that only methodologically sound studies should be included in a meta-analysis, a practice called 'best evidence synthesis'.[61] Other meta-analysts would include weaker studies, and add a study-level predictor variable that reflects the methodological quality of the studies to examine the effect of study quality on the effect size.[62] However, others have argued that a better approach is to preserve information about the variance in the study sample, casting as wide a net as possible, and that methodological selection criteria introduce unwanted subjectivity, defeating the purpose of the approach.[63]
Publication bias: the file drawer problem
Another potential pitfall is the reliance on the available body of published studies, which may create exaggerated outcomes due to publication bias, as studies which show negative results or insignificant results are less likely to be published. For example, pharmaceutical companies have been known to hide negative studies and researchers may have overlooked unpublished studies such as dissertation studies or conference abstracts that did not reach publication. This is not easily solved, as one cannot know how many studies have gone unreported.[64]
This file drawer problem (characterized by negative or non-significant results being tucked away in a cabinet), can result in a biased distribution of effect sizes thus creating a serious base rate fallacy, in which the significance of the published studies is overestimated, as other studies were either not submitted for publication or were rejected. This should be seriously considered when interpreting the outcomes of a meta-analysis.[64][65]
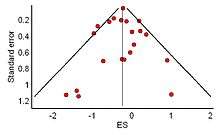
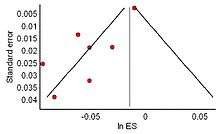
The distribution of effect sizes can be visualized with a funnel plot which (in its most common version) is a scatter plot of standard error versus the effect size. It makes use of the fact that the smaller studies (thus larger standard errors) have more scatter of the magnitude of effect (being less precise) while the larger studies have less scatter and form the tip of the funnel. If many negative studies were not published, the remaining positive studies give rise to a funnel plot in which the base is skewed to one side (asymmetry of the funnel plot). In contrast, when there is no publication bias, the effect of the smaller studies has no reason to be skewed to one side and so a symmetric funnel plot results. This also means that if no publication bias is present, there would be no relationship between standard error and effect size.[66] A negative or positive relation between standard error and effect size would imply that smaller studies that found effects in one direction only were more likely to be published and/or to be submitted for publication.
Apart from the visual funnel plot, statistical methods for detecting publication bias have also been proposed. These are controversial because they typically have low power for detection of bias, but also may make false positives under some circumstances.[67] For instance small study effects (biased smaller studies), wherein methodological differences between smaller and larger studies exist, may cause asymmetry in effect sizes that resembles publication bias. However, small study effects may be just as problematic for the interpretation of meta-analyses, and the imperative is on meta-analytic authors to investigate potential sources of bias.
A Tandem Method for analyzing publication bias has been suggested for cutting down false positive error problems.[68] This Tandem method consists of three stages. Firstly, one calculates Orwin's fail-safe N, to check how many studies should be added in order to reduce the test statistic to a trivial size. If this number of studies is larger than the number of studies used in the meta-analysis, it is a sign that there is no publication bias, as in that case, one needs a lot of studies to reduce the effect size. Secondly, one can do an Egger's regression test, which tests whether the funnel plot is symmetrical. As mentioned before: a symmetrical funnel plot is a sign that there is no publication bias, as the effect size and sample size are not dependent. Thirdly, one can do the trim-and-fill method, which imputes data if the funnel plot is asymmetrical.
The problem of publication bias is not trivial as it is suggested that 25% of meta-analyses in the psychological sciences may have suffered from publication bias.[68] However, low power of existing tests and problems with the visual appearance of the funnel plot remain an issue, and estimates of publication bias may remain lower than what truly exists.
Most discussions of publication bias focus on journal practices favoring publication of statistically significant findings. However, questionable research practices, such as reworking statistical models until significance is achieved, may also favor statistically significant findings in support of researchers' hypotheses.[69][70]
Problems related to studies not reporting non-statistically significant effects
Studies often do not report the effects when they do not reach statistical significance. For example, they may simply say that the groups did not show statistically significant differences, without report any other information (e.g. a statistic or p-value). Exclusion of these studies would lead to a situation similar to publication bias, but their inclusion (assuming null effects) would also bias the meta-analysis. MetaNSUE, a method created by Joaquim Radua, has shown to allow researchers to include unbiasedly these studies.[71] Its steps are as follows:
- Maximum likelihood estimation of the meta-analytic effect and the heterogeneity between studies.
- Multiple imputation of the NSUEs adding noise to the estimate of the effect.
- Separate meta-analyses for each imputed dataset.
- Pooling of the results of these meta-analyses.
Problems related to the statistical approach
Other weaknesses are that it has not been determined if the statistically most accurate method for combining results is the fixed, IVhet, random or quality effect models, though the criticism against the random effects model is mounting because of the perception that the new random effects (used in meta-analysis) are essentially formal devices to facilitate smoothing or shrinkage and prediction may be impossible or ill-advised.[72] The main problem with the random effects approach is that it uses the classic statistical thought of generating a "compromise estimator" that makes the weights close to the naturally weighted estimator if heterogeneity across studies is large but close to the inverse variance weighted estimator if the between study heterogeneity is small. However, what has been ignored is the distinction between the model we choose to analyze a given dataset, and the mechanism by which the data came into being.[73] A random effect can be present in either of these roles, but the two roles are quite distinct. There's no reason to think the analysis model and data-generation mechanism (model) are similar in form, but many sub-fields of statistics have developed the habit of assuming, for theory and simulations, that the data-generation mechanism (model) is identical to the analysis model we choose (or would like others to choose). As a hypothesized mechanisms for producing the data, the random effect model for meta-analysis is silly and it is more appropriate to think of this model as a superficial description and something we choose as an analytical tool – but this choice for meta-analysis may not work because the study effects are a fixed feature of the respective meta-analysis and the probability distribution is only a descriptive tool.[73]
Problems arising from agenda-driven bias
The most severe fault in meta-analysis[74] often occurs when the person or persons doing the meta-analysis have an economic, social, or political agenda such as the passage or defeat of legislation. People with these types of agendas may be more likely to abuse meta-analysis due to personal bias. For example, researchers favorable to the author's agenda are likely to have their studies cherry-picked while those not favorable will be ignored or labeled as "not credible". In addition, the favored authors may themselves be biased or paid to produce results that support their overall political, social, or economic goals in ways such as selecting small favorable data sets and not incorporating larger unfavorable data sets. The influence of such biases on the results of a meta-analysis is possible because the methodology of meta-analysis is highly malleable.[75]
A 2011 study done to disclose possible conflicts of interests in underlying research studies used for medical meta-analyses reviewed 29 meta-analyses and found that conflicts of interests in the studies underlying the meta-analyses were rarely disclosed. The 29 meta-analyses included 11 from general medicine journals, 15 from specialty medicine journals, and three from the Cochrane Database of Systematic Reviews. The 29 meta-analyses reviewed a total of 509 randomized controlled trials (RCTs). Of these, 318 RCTs reported funding sources, with 219 (69%) receiving funding from industry (i.e. one or more authors having financial ties to the pharmaceutical industry). Of the 509 RCTs, 132 reported author conflict of interest disclosures, with 91 studies (69%) disclosing one or more authors having financial ties to industry. The information was, however, seldom reflected in the meta-analyses. Only two (7%) reported RCT funding sources and none reported RCT author-industry ties. The authors concluded "without acknowledgment of COI due to industry funding or author industry financial ties from RCTs included in meta-analyses, readers' understanding and appraisal of the evidence from the meta-analysis may be compromised."[76]
For example, in 1998, a US federal judge found that the United States Environmental Protection Agency had abused the meta-analysis process to produce a study claiming cancer risks to non-smokers from environmental tobacco smoke (ETS) with the intent to influence policy makers to pass smoke-free–workplace laws. The judge found that:
EPA's study selection is disturbing. First, there is evidence in the record supporting the accusation that EPA "cherry picked" its data. Without criteria for pooling studies into a meta-analysis, the court cannot determine whether the exclusion of studies likely to disprove EPA's a priori hypothesis was coincidence or intentional. Second, EPA's excluding nearly half of the available studies directly conflicts with EPA's purported purpose for analyzing the epidemiological studies and conflicts with EPA's Risk Assessment Guidelines. See ETS Risk Assessment at 4-29 ("These data should also be examined in the interest of weighing all the available evidence, as recommended by EPA's carcinogen risk assessment guidelines (U.S. EPA, 1986a) (emphasis added)). Third, EPA's selective use of data conflicts with the Radon Research Act. The Act states EPA's program shall "gather data and information on all aspects of indoor air quality" (Radon Research Act § 403(a)(1)) (emphasis added).[77]
As a result of the abuse, the court vacated Chapters 1–6 of and the Appendices to EPA's "Respiratory Health Effects of Passive Smoking: Lung Cancer and other Disorders".[77]
Applications in modern science
Modern statistical meta-analysis does more than just combine the effect sizes of a set of studies using a weighted average. It can test if the outcomes of studies show more variation than the variation that is expected because of the sampling of different numbers of research participants. Additionally, study characteristics such as measurement instrument used, population sampled, or aspects of the studies' design can be coded and used to reduce variance of the estimator (see statistical models above). Thus some methodological weaknesses in studies can be corrected statistically. Other uses of meta-analytic methods include the development and validation of clinical prediction models, where meta-analysis may be used to combine individual participant data from different research centers and to assess the model's generalisability,[78][79] or even to aggregate existing prediction models.[80]
Meta-analysis can be done with single-subject design as well as group research designs. This is important because much research has been done with single-subject research designs. Considerable dispute exists for the most appropriate meta-analytic technique for single subject research.[81]
Meta-analysis leads to a shift of emphasis from single studies to multiple studies. It emphasizes the practical importance of the effect size instead of the statistical significance of individual studies. This shift in thinking has been termed "meta-analytic thinking". The results of a meta-analysis are often shown in a forest plot.
Results from studies are combined using different approaches. One approach frequently used in meta-analysis in health care research is termed 'inverse variance method'. The average effect size across all studies is computed as a weighted mean, whereby the weights are equal to the inverse variance of each study's effect estimator. Larger studies and studies with less random variation are given greater weight than smaller studies. Other common approaches include the Mantel–Haenszel method[82] and the Peto method.[83]
Seed-based d mapping (formerly signed differential mapping, SDM) is a statistical technique for meta-analyzing studies on differences in brain activity or structure which used neuroimaging techniques such as fMRI, VBM or PET.
Different high throughput techniques such as microarrays have been used to understand Gene expression. MicroRNA expression profiles have been used to identify differentially expressed microRNAs in particular cell or tissue type or disease conditions or to check the effect of a treatment. A meta-analysis of such expression profiles was performed to derive novel conclusions and to validate the known findings.[84]
See also
References
- Greenland S, O' Rourke K: Meta-Analysis. Page 652 in Modern Epidemiology, 3rd ed. Edited by Rothman KJ, Greenland S, Lash T. Lippincott Williams and Wilkins; 2008.
- Walker E, Hernandez AV, Kattan MW (2008). "Meta-analysis: Its strengths and limitations". Cleve Clin J Med. 75 (6): 431–9. doi:10.3949/ccjm.75.6.431. PMID 18595551.
- Wanous, John P.; Sullivan, Sherry E.; Malinak, Joyce (1989). "The role of judgment calls in meta-analysis". Journal of Applied Psychology. 74 (2): 259–264. doi:10.1037/0021-9010.74.2.259. ISSN 0021-9010.
- "Glossary at Cochrane Collaboration". cochrane.org.
- Gravetter, Frederick J.; Forzano, Lori-Ann B. (1 January 2018). Research Methods for the Behavioral Sciences. Cengage Learning. p. 36. ISBN 9781337613316.
Some examples of secondary sources are (1) books and textbooks in which the author describes and summarizes past research, (2) review articles or meta-analyses...
- Adams, Kathrynn A.; Lawrence, Eva K. (2 February 2018). Research Methods, Statistics, and Applications. SAGE Publications. ISBN 9781506350462.
The most common types of secondary sources found in academic journals are literature reviews and meta-analyses.
- PLACKETT, R. L. (1958). "Studies in the History of Probability and Statistics: Vii. The Principle of the Arithmetic Mean". Biometrika. 45 (1–2): 133. doi:10.1093/biomet/45.1-2.130. Retrieved 29 May 2016.
- Pearson K (1904). "Report on certain enteric fever inoculation statistics". BMJ. 2 (2288): 1243–1246. doi:10.1136/bmj.2.2288.1243. PMC 2355479. PMID 20761760.
- Nordmann AJ, Kasenda B, Briel M (9 March 2012). "Meta-analyses: what they can and cannot do". Swiss Medical Weekly. 142: w13518. doi:10.4414/smw.2012.13518. PMID 22407741.
- O'Rourke K (1 December 2007). "An historical perspective on meta-analysis: dealing quantitatively with varying study results". J R Soc Med. 100 (12): 579–582. doi:10.1258/jrsm.100.12.579. PMC 2121629. PMID 18065712.
- Pratt JG, Rhine JB, Smith BM, Stuart CE, Greenwood JA. Extra-Sensory Perception after Sixty Years: A Critical Appraisal of the Research in Extra-Sensory Perception. New York: Henry Holt, 1940
- Glass G. V (1976). "Primary, secondary, and meta-analysis of research". Educational Researcher. 5 (10): 3–8. doi:10.3102/0013189X005010003.
- Cochran WG (1937). "Problems Arising in the Analysis of a Series of Similar Experiments". Journal of the Royal Statistical Society. 4 (1): 102–118. doi:10.2307/2984123. JSTOR 2984123.
- Cochran WG, Carroll SP (1953). "A Sampling Investigation of the Efficiency of Weighting Inversely as the Estimated Variance". Biometrics. 9 (4): 447–459. doi:10.2307/3001436. JSTOR 3001436.
- "The PRISMA statement". Prisma-statement.org. 2 February 2012. Retrieved 2 February 2012.
- Debray, Thomas P. A.; Moons, Karel G. M.; van Valkenhoef, Gert; Efthimiou, Orestis; Hummel, Noemi; Groenwold, Rolf H. H.; Reitsma, Johannes B.; on behalf of the GetReal methods review group (1 December 2015). "Get real in individual participant data (IPD) meta-analysis: a review of the methodology". Research Synthesis Methods. 6 (4): 293–309. doi:10.1002/jrsm.1160. ISSN 1759-2887. PMC 5042043. PMID 26287812.
- Debray TP, Moons KG, Abo-Zaid GM, Koffijberg H, Riley RD (2013). "Individual participant data meta-analysis for a binary outcome: one-stage or two-stage?". PLOS One. 8 (4): e60650. Bibcode:2013PLoSO...860650D. doi:10.1371/journal.pone.0060650. PMC 3621872. PMID 23585842.
- Burke, Danielle L.; Ensor, Joie; Riley, Richard D. (28 February 2017). "Meta-analysis using individual participant data: one-stage and two-stage approaches, and why they may differ". Statistics in Medicine. 36 (5): 855–875. doi:10.1002/sim.7141. ISSN 1097-0258. PMC 5297998. PMID 27747915.
- Helfenstein U (2002). "Data and models determine treatment proposals--an illustration from meta-analysis". Postgrad Med J. 78 (917): 131–4. doi:10.1136/pmj.78.917.131. PMC 1742301. PMID 11884693.
- Senn S (2007). "Trying to be precise about vagueness". Stat Med. 26 (7): 1417–30. doi:10.1002/sim.2639. PMID 16906552.
- Al Khalaf MM, Thalib L, Doi SA (2011). "Combining heterogenous studies using the random-effects model is a mistake and leads to inconclusive meta-analyses". Journal of Clinical Epidemiology. 64 (2): 119–23. doi:10.1016/j.jclinepi.2010.01.009. PMID 20409685.
- Brockwell S.E.; Gordon I.R. (2001). "A comparison of statistical methods for meta-analysis". Statistics in Medicine. 20 (6): 825–840. doi:10.1002/sim.650. PMID 11252006.
- Noma H (December 2011). "Confidence intervals for a random-effects meta-analysis based on Bartlett-type corrections". Stat Med. 30 (28): 3304–12. doi:10.1002/sim.4350. hdl:2433/152046. PMID 21964669.
- Brockwell SE, Gordon IR (2007). "A simple method for inference on an overall effect in meta-analysis". Statistics in Medicine. 26 (25): 4531–4543. doi:10.1002/sim.2883. PMID 17397112.
- Sidik K, Jonkman JN (2002). "A simple confidence interval for meta-analysis". Statistics in Medicine. 21 (21): 3153–3159. doi:10.1002/sim.1262. PMID 12375296.
- Jackson D, Bowden J (2009). "A re-evaluation of the 'quantile approximation method' for random effects meta-analysis". Stat Med. 28 (2): 338–48. doi:10.1002/sim.3487. PMC 2991773. PMID 19016302.
- Poole C, Greenland S (September 1999). "Random-effects meta-analyses are not always conservative". Am J Epidemiol. 150 (5): 469–75. doi:10.1093/oxfordjournals.aje.a010035. PMID 10472946.
- Riley RD, Higgins JP, Deeks JJ (2011). "Interpretation of random effects meta-analyses". British Medical Journal. 342: d549. doi:10.1136/bmj.d549. PMID 21310794.
- Kriston L (2013). "Dealing with clinical heterogeneity in meta-analysis. Assumptions, methods, interpretation". Int J Methods Psychiatr Res. 22 (1): 1–15. doi:10.1002/mpr.1377. PMC 6878481. PMID 23494781.
- DerSimonian R, Laird N (1986). "Meta-analysis in clinical trials". Control Clin Trials. 7 (3): 177–88. doi:10.1016/0197-2456(86)90046-2. PMID 3802833.
- Kontopantelis, Evangelos; Reeves, David (1 August 2010). "Metaan: Random-effects meta-analysis". Stata Journal. 10 (3): 395–407. doi:10.1177/1536867X1001000307 – via ResearchGate.
- Kontopantelis, Evangelos; Reeves, David (2009). "MetaEasy:A Meta-Analysis Add-In for Microsoft Excel, Journal of Statistical Software 2009". Journal of Statistical Software. 30 (7). doi:10.18637/jss.v030.i07.
- "Developer's website". Statanalysis.co.uk. Retrieved 18 September 2018.
- Kontopantelis E, Reeves D (2012). "Performance of statistical methods for meta-analysis when true study effects are non-normally distributed: A simulation study". Statistical Methods in Medical Research. 21 (4): 409–26. doi:10.1177/0962280210392008. PMID 21148194.
- Kontopantelis E, Reeves D (2012). "Performance of statistical methods for meta-analysis when true study effects are non-normally distributed: a comparison between DerSimonian-Laird and restricted maximum likelihood". SMMR. 21 (6): 657–9. doi:10.1177/0962280211413451. PMID 23171971.
- Kontopantelis E, Springate DA, Reeves D (2013). Friede T (ed.). "A Re-Analysis of the Cochrane Library Data: The Dangers of Unobserved Heterogeneity in Meta-Analyses". PLOS One. 8 (7): e69930. Bibcode:2013PLoSO...869930K. doi:10.1371/journal.pone.0069930. PMC 3724681. PMID 23922860.
- Kontopantelis, Evangelos; Reeves, David (27 September 2013). "A short guide and a forest plot command (ipdforest) for one-stage meta-analysis". Stata Journal. 13 (3): 574–587. doi:10.1177/1536867X1301300308 – via ResearchGate.
- "MetaXL User Guide" (PDF). Retrieved 18 September 2018.
- "MetaXL software page". Epigear.com. 3 June 2017. Retrieved 18 September 2018.
- Doi SA, Barendregt JJ, Khan S, Thalib L, Williams GM (2015). "Advances in the Meta-analysis of heterogeneous clinical trials I: The inverse variance heterogeneity model" (PDF). Contemp Clin Trials. 45 (Pt A): 130–8. doi:10.1016/j.cct.2015.05.009. PMID 26003435.
- Doi SA, Thalib L (2008). "A quality-effects model for meta-analysis". Epidemiology. 19 (1): 94–100. doi:10.1097/EDE.0b013e31815c24e7. PMID 18090860.
- Doi SA, Barendregt JJ, Mozurkewich EL (2011). "Meta-analysis of heterogeneous clinical trials: an empirical example". Contemp Clin Trials. 32 (2): 288–98. doi:10.1016/j.cct.2010.12.006. PMID 21147265.
- Doi SA, Barendregt JJ, Williams GM, Khan S, Thalib L (2015). "Simulation Comparison of the Quality Effects and Random Effects Methods of Meta-analysis". Epidemiology. 26 (4): e42–4. doi:10.1097/EDE.0000000000000289. PMID 25872162.
- Doi SA, Barendregt JJ, Khan S, Thalib L, Williams GM (2015). "Advances in the meta-analysis of heterogeneous clinical trials II: The quality effects model". Contemp Clin Trials. 45 (Pt A): 123–9. doi:10.1016/j.cct.2015.05.010. PMID 26003432.
- Bucher H. C.; Guyatt G. H.; Griffith L. E.; Walter S. D. (1997). "The results of direct and indirect treatment comparisons in meta-analysis of randomized controlled trials". J Clin Epidemiol. 50 (6): 683–691. doi:10.1016/s0895-4356(97)00049-8. PMID 9250266.
- Valkenhoef G.; Lu G.; Brock B.; Hillege H.; Ades A. E.; Welton N. J. (2012). "Automating network meta‐analysis". Research Synthesis Methods. 3 (4): 285–299. doi:10.1002/jrsm.1054. PMID 26053422.
- Brooks SP, Gelman A (1998). "General methods for monitoring convergence of iterative simulations" (PDF). Journal of Computational and Graphical Statistics. 7 (4): 434–455. doi:10.1080/10618600.1998.10474787.
- van Valkenhoef G, Lu G, de Brock B, Hillege H, Ades AE, Welton NJ. Automating network meta-analysis. Res Synth Methods. 2012 Dec;3(4):285-99.
- Senn S, Gavini F, Magrez D, Scheen A (April 2013). "Issues in performing a network meta-analysis". Stat Methods Med Res. 22 (2): 169–89. doi:10.1177/0962280211432220. PMID 22218368.
- White IR (2011). "Multivariate random-effects meta-regression: updates to mvmeta". The Stata Journal. 11 (2): 255–270. doi:10.1177/1536867X1101100206.
- van Valkenhoef G, Lu G, de Brock B, Hillege H, Ades AE, Welton NJ. Automating network meta-analysis. Res Synth Methods. 2012 Dec;3(4):285-99
- Willis BH, Hyde CJ (2014). "Estimating a test's accuracy using tailored meta-analysis – How setting-specific data may aid study selection". Journal of Clinical Epidemiology. 67 (5): 538–546. doi:10.1016/j.jclinepi.2013.10.016. PMID 24447592.
- Willis BH, Hyde CJ (2015). "What is the test's accuracy in my practice population? Tailored meta-analysis provides a plausible estimate". Journal of Clinical Epidemiology. 68 (8): 847–854. doi:10.1016/j.jclinepi.2014.10.002. PMID 25479685.
- Zhang H, Deng L, Schiffman M, Qin J, Yu K (2020). "Generalized integration model for improved statistical inference by leveraging external summary data". Biometrika. doi:10.1093/biomet/asaa014.
- Higgins JP, Altman DG, Gøtzsche PC, Jüni P, Moher D, Oxman AD, Savovic J, Schulz KF, Weeks L, Sterne JA; Cochrane Bias Methods Group; Cochrane Statistical Methods Group (2011). "The Cochrane Collaboration's tool for assessing risk of bias in randomised trials". BMJ. 343: d5928. doi:10.1136/bmj.d5928. PMC 3196245. PMID 22008217.CS1 maint: multiple names: authors list (link)
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, QUADAS-2 Group (2011). "QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies". Annals of Internal Medicine. 155 (8): 529–36. doi:10.7326/0003-4819-155-8-201110180-00009. PMID 22007046.CS1 maint: multiple names: authors list (link)
- Royston P, Parmar MK, Sylvester R (2004). "Construction and validation of a prognostic model across several studies, with an application in superficial bladder cancer". Statistics in Medicine. 23 (6): 907–26. doi:10.1002/sim.1691. PMID 15027080.
- Willis BH, Riley RD (2017). "Measuring the statistical validity of summary meta-analysis and meta-regression results for use in clinical practice". Statistics in Medicine. 36 (21): 3283–3301. doi:10.1002/sim.7372. PMC 5575530. PMID 28620945.
- Riley RD, Ahmed I, Debray TP, Willis BH, Noordzij P, Higgins JP, Deeks JJ (2015). "Summarising and validating test accuracy results across multiple studies for use in clinical practice". Statistics in Medicine. 34 (13): 2081–2103. doi:10.1002/sim.6471. PMC 4973708. PMID 25800943.
- LeLorier J, Grégoire G, Benhaddad A, Lapierre J, Derderian F (1997). "Discrepancies between Meta-Analyses and Subsequent Large Randomized, Controlled Trials". New England Journal of Medicine. 337 (8): 536–542. doi:10.1056/NEJM199708213370806. PMID 9262498.
- Slavin RE (1986). "Best-Evidence Synthesis: An Alternative to Meta-Analytic and Traditional Reviews". Educational Researcher. 15 (9): 5–9. doi:10.3102/0013189X015009005.
- Hunter, Schmidt, & Jackson, John E. (1982). Meta-analysis: Cumulating research findings across studies. Beverly Hills, California: Sage.CS1 maint: multiple names: authors list (link)
- Glass, McGaw, & Smith (1981). Meta-analysis in social research. Beverly Hills, CA: Sage.CS1 maint: multiple names: authors list (link)
- Rosenthal R (1979). "The "File Drawer Problem" and the Tolerance for Null Results". Psychological Bulletin. 86 (3): 638–641. doi:10.1037/0033-2909.86.3.638.
- Hunter, John E; Schmidt, Frank L (1990). Methods of Meta-Analysis: Correcting Error and Bias in Research Findings. Newbury Park, California; London; New Delhi: SAGE Publications.
- Light & Pillemer (1984). Summing up: The science of reviewing research. Cambridge, CA: Harvard University Pree.
- Ioannidis JP, Trikalinos TA (2007). "The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey". CMAJ. 176 (8): 1091–6. doi:10.1503/cmaj.060410. PMC 1839799. PMID 17420491.
- Ferguson CJ, Brannick MT (2012). "Publication bias in psychological science: prevalence, methods for identifying and controlling, and implications for the use of meta-analyses". Psychol Methods. 17 (1): 120–8. doi:10.1037/a0024445. PMID 21787082.
- Simmons JP, Nelson LD, Simonsohn U (2011). "False-positive psychology: undisclosed flexibility in data collection and analysis allows presenting anything as significant". Psychol Sci. 22 (11): 1359–66. doi:10.1177/0956797611417632. PMID 22006061.
- LeBel, E.; Peters, K. (2011). "Fearing the future of empirical psychology: Bem's (2011) evidence of psi as a case study of deficiencies in modal research practice" (PDF). Review of General Psychology. 15 (4): 371–379. doi:10.1037/a0025172. Archived from the original (PDF) on 24 November 2012.
- Radua, J.; Schmidt, A.; Borgwardt, S.; Heinz, A.; Schlagenhauf, F.; McGuire, P.; Fusar-Poli, P. (2015). "Ventral Striatal Activation During Reward Processing in Psychosis: A Neurofunctional Meta-Analysis". JAMA Psychiatry. 72 (12): 1243–1251. doi:10.1001/jamapsychiatry.2015.2196. PMID 26558708.
- Hodges, Jim, and Clayton, Murray K. Random Effects: Old and New. Statistical Science XX: XX–XX. URL http://www Archived 24 May 2011 at the Wayback Machine. biostat. umn. edu/~ hodges/Hodges-ClaytonREONsubToStatSci (2011)
- Hodges JS. Random effects old and new. In Hodges JS. Richly parameterized linear models: additive, time series, and spatial models using random effects. USA: CRC Press, 2013: 285–302.
- H. Sabhan
- Stegenga J (2011). "Is meta-analysis the platinum standard of evidence?". Stud Hist Philos Biol Biomed Sci. 42 (4): 497–507. doi:10.1016/j.shpsc.2011.07.003. PMID 22035723.
- Roseman M, Milette K, Bero LA, Coyne JC, Lexchin J, Turner EH, Thombs BD (2011), "Reporting of Conflicts of Interest in Meta-analyses of Trials of Pharmacological Treatments", Journal of the American Medical Association, 305 (10): 1008–1017, doi:10.1001/jama.2011.257, PMID 21386079CS1 maint: multiple names: authors list (link)
- "The Osteen Decision". The United States District Court for the Middle District of North Carolina. 17 July 1998. Retrieved 18 March 2017.
- Debray, Thomas P. A.; Riley, Richard D.; Rovers, Maroeska M.; Reitsma, Johannes B.; Moons, Karel G. M. (13 October 2015). "Individual Participant Data (IPD) Meta-analyses of Diagnostic and Prognostic Modeling Studies: Guidance on Their Use". PLOS Medicine. 12 (10): e1001886. doi:10.1371/journal.pmed.1001886. PMC 4603958. PMID 26461078.
- Debray TP, Moons KG, Ahmed I, Koffijberg H, Riley RD (2013). "A framework for developing, implementing, and evaluating clinical prediction models in an individual participant data meta-analysis". Statistics in Medicine. 32 (18): 3158–80. doi:10.1002/sim.5732. PMID 23307585.
- Debray TP, Koffijberg H, Vergouwe Y, Moons KG, Steyerberg EW (2012). "Aggregating published prediction models with individual participant data: a comparison of different approaches". Statistics in Medicine. 31 (23): 2697–2712. doi:10.1002/sim.5412. PMID 22733546.
- Van den Noortgate W, Onghena P (2007). "Aggregating Single-Case Results". The Behavior Analyst Today. 8 (2): 196–209. doi:10.1037/h0100613.
- Mantel N, Haenszel W (1959). "Statistical aspects of the analysis of data from the retrospective analysis of disease". Journal of the National Cancer Institute. 22 (4): 719–748. doi:10.1093/jnci/22.4.719. PMID 13655060.
- "9.4.4.2 Peto odds ratio method". Cochrane Handbook for Systematic Reviews of Interventions v 5.1.0. March 2011.
- Bargaje R, Hariharan M, Scaria V, Pillai B (2010). "Consensus miRNA expression profiles derived from interplatform normalization of microarray data". RNA. 16 (1): 16–25. doi:10.1261/rna.1688110. PMC 2802026. PMID 19948767.
Further reading
- Cooper, H. & Hedges, L.V. (1994). The Handbook of Research Synthesis. New York: Russell Sage.
- Cornell, J. E. & Mulrow, C. D. (1999). Meta-analysis. In: H. J. Adèr & G. J. Mellenbergh (Eds). Research Methodology in the social, behavioral and life sciences (pp. 285–323). London: Sage.
- Normand SL (1999). "Tutorial in Biostatistics. Meta-Analysis: Formulating, Evaluating, Combining, and Reporting". Statistics in Medicine. 18 (3): 321–359. doi:10.1002/(SICI)1097-0258(19990215)18:3<321::AID-SIM28>3.0.CO;2-P. PMID 10070677.
- Sutton, A.J., Jones, D.R., Abrams, K.R., Sheldon, T.A., & Song, F. (2000). Methods for Meta-analysis in Medical Research. London: John Wiley. ISBN 0-471-49066-0
- Higgins JPT, Green S (editors). Cochrane Handbook for Systematic Reviews of Interventions Version 5.0.1 [updated September 2008]. The Cochrane Collaboration, 2008. Available from www.cochrane-handbook.org
- Thompson SG, Pocock SJ; Pocock, Stuart J (2 November 1991). "Can meta-analysis be trusted?" (PDF). The Lancet. 338 (8775): 1127–1130. doi:10.1016/0140-6736(91)91975-Z. PMID 1682553. Archived from the original (PDF) on 22 November 2011. Retrieved 17 June 2011.. Explores two contrasting views: does meta-analysis provide "objective, quantitative methods for combining evidence from separate but similar studies" or merely "statistical tricks which make unjustified assumptions in producing oversimplified generalisations out of a complex of disparate studies"?
- Wilson, D. B., & Lipsey, M. W. (2001). Practical meta-analysis. Thousand Oaks: Sage publications. ISBN 0-7619-2168-0
- O'Rourke, K. (2007) Just the history from the combining of information: investigating and synthesizing what is possibly common in clinical observations or studies via likelihood. Oxford: University of Oxford, Department of Statistics. Gives technical background material and details on the "An historical perspective on meta-analysis" paper cited in the references.
- Owen, A. B. (2009). "Karl Pearson's meta-analysis revisited". Annals of Statistics, 37 (6B), 3867–3892. Supplementary report.
- Ellis, Paul D. (2010). The Essential Guide to Effect Sizes: An Introduction to Statistical Power, Meta-Analysis and the Interpretation of Research Results. United Kingdom: Cambridge University Press. ISBN 0-521-14246-6
- Bonett DG, Price RM (2015). "Varying coefficient meta-analysis methods for odds ratios and risk ratios". Psychol Methods. 20 (3): 394–406. doi:10.1037/met0000032. PMID 25751513.
- Bonett DG, Price RM (2014). "Meta-analysis methods for risk differences". Br J Math Stat Psychol. 67 (3): 371–87. doi:10.1111/bmsp.12024. PMID 23962020.
- Bonett DG (2010). "Varying coefficient meta-analytic methods for alpha reliability". Psychol Methods. 15 (4): 368–85. doi:10.1037/a0020142. PMID 20853952.
- Bonett DG (2009). "Meta-analytic interval estimation for standardized and unstandardized mean differences". Psychol Methods. 14 (3): 225–38. doi:10.1037/a0016619. PMID 19719359.
- Bonett DG (2008). "Meta-analytic interval estimation for bivariate correlations". Psychol Methods. 13 (3): 173–81. doi:10.1037/a0012868. PMID 18778150.
External links
| Wikiversity has learning resources about Meta-analysis |
- Cochrane Handbook for Systematic Reviews of Interventions
- Meta-Analysis at 25 (Gene V Glass)
- Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement, "an evidence-based minimum set of items for reporting in systematic reviews and meta-analyses."
- “metansue” R package and graphical interface