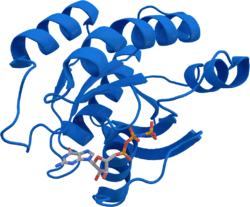
KRAS
The KRAS gene provides instructions for making a protein called K-Ras, part of the RAS/MAPK pathway. The protein relays signals from outside the cell to the cell's nucleus. These signals instruct the cell to grow and divide (proliferate) or to mature and take on specialized functions (differentiate). The K-Ras protein is a GTPase, which means it converts a molecule called GTP into another molecule called GDP. In this way the K-Ras protein acts like a switch that is turned on and off by the GTP and GDP molecules. To transmit signals, it must be turned on by attaching (binding) to a molecule of GTP. The K-Ras protein is turned off (inactivated) when it converts the GTP to GDP. When the protein is bound to GDP, it does not relay signals to the cell's nucleus. It is called KRAS because it was first identified as an oncogene in Kirsten RAt Sarcoma virus.[5] The viral oncogene was derived from cellular genome. Thus, KRAS gene in cellular genome is called a proto-oncogene.
The gene product was first found as a p21 GTPase.[6][7] Like other members of the ras subfamily, the KRAS protein is a GTPase and is an early player in many signal transduction pathways. KRAS is usually tethered to cell membranes because of the presence of an isoprene group on its C-terminus. There are two protein products of the KRAS gene in mammalian cells that result from the use of alternative exon 4 (exon 4A and 4B respectively): K-Ras4A and K-Ras4B, these proteins have different structure in their C-terminal region and use different mechanisms to localize to cellular membranes including the plasma membrane.[8]
Function
KRAS acts as a molecular on/off switch, using protein dynamics. Once it is allosterically activated, it recruits and activates proteins necessary for the propagation of growth factors, as well as other cell signaling receptors like c-Raf and PI 3-kinase. KRAS upregulates the GLUT1 glucose transporter, thereby contributing to the Warburg effect in cancer cells.[9] KRAS binds to GTP in its active state. It also possesses an intrinsic enzymatic activity which cleaves the terminal phosphate of the nucleotide, converting it to GDP. Upon conversion of GTP to GDP, KRAS is deactivated. The rate of conversion is usually slow, but can be increased dramatically by an accessory protein of the GTPase-activating protein (GAP) class, for example RasGAP. In turn, KRAS can bind to proteins of the Guanine Nucleotide Exchange Factor (GEF) class (such as SOS1), which forces the release of bound nucleotide (GDP). Subsequently, KRAS binds GTP present in the cytosol and the GEF is released from ras-GTP.
Other members of the Ras family include: HRAS and NRAS. These proteins all are regulated in the same manner and appear to differ in their sites of action within the cell.
Clinical significance when mutated
This proto-oncogene is a Kirsten ras oncogene homolog from the mammalian ras gene family. A single amino acid substitution, and in particular a single nucleotide substitution, is responsible for an activating mutation. The transforming protein that results is implicated in various malignancies, including lung adenocarcinoma,[10] mucinous adenoma, ductal carcinoma of the pancreas and colorectal cancer.[11][12]
Several germline KRAS mutations have been found to be associated with Noonan syndrome[13] and cardio-facio-cutaneous syndrome.[14]
Somatic KRAS mutations are found at high rates in leukemias, colorectal cancer,[15] pancreatic cancer[16] and lung cancer.[17]
Colorectal cancer
The impact of KRAS mutations is heavily dependent on the order of mutations. Primary KRAS mutations generally lead to a self-limiting hyperplastic or borderline lesion, but if they occur after a previous APC mutation it often progresses to cancer.[18] KRAS mutations are more commonly observed in cecal cancers than colorectal cancers located in any other places from ascending colon to rectum.[19][20]
KRAS mutation is predictive of a very poor response to panitumumab (Vectibix) and cetuximab (Erbitux) therapy in colorectal cancer.[21]
Currently, the most reliable way to predict whether a colorectal cancer patient will respond to one of the EGFR-inhibiting drugs is to test for certain “activating” mutations in the gene that encodes KRAS, which occurs in 30%–50% of colorectal cancers. Studies show patients whose tumors express the mutated version of the KRAS gene will not respond to cetuximab or panitumumab.[22]
Although presence of the wild-type (or normal) KRAS gene does not guarantee that these drugs will work, a number of large studies[23][24] have shown that cetuximab has significant efficacy in mCRC patients with KRAS wild-type tumors. In the Phase III CRYSTAL study, published in 2009, patients with the wild-type KRAS gene treated with Erbitux plus chemotherapy showed a response rate of up to 59% compared to those treated with chemotherapy alone. Patients with the KRAS wild-type gene also showed a 32% decreased risk of disease progression compared to patients receiving chemotherapy alone.[24]
Emergence of KRAS mutations is a frequent driver of acquired resistance to cetuximab anti-EGFR therapy in colorectal cancers. The emergence of KRAS mutant clones can be detected non-invasively months before radiographic progression. It suggests to perform an early initiation of a MEK inhibitor as a rational strategy for delaying or reversing drug resistance.[25]
KRAS amplification
KRAS gene can also be amplified in colorectal cancer. KRAS amplification is mutually exclusive with KRAS mutations. Tumors or cell lines harboring this genetic lesion are not responsive to EGFR inhibitors. Although KRAS amplification is an infrequent event in colorectal cancer, it might be responsible for precluding response to anti-EGFR treatment in some patients.[26] Amplification of wild-type Kras has also been observed in ovarian,[27] gastric, uterine, and lung cancers.[28]
Lung cancer
Whether a patient is positive or negative for a mutation in the epidermal growth factor receptor (EGFR) will predict how patients will respond to certain EGFR antagonists such as erlotinib (Tarceva) or gefitinib (Iressa). Patients who harbor an EGFR mutation have a 60% response rate to erlotinib. However, the mutation of KRAS and EGFR are generally mutually exclusive.[29][30][31] Lung cancer patients who are positive for KRAS mutation (and the EGFR status would be wild type) have a low response rate to erlotinib or gefitinib estimated at 5% or less.[29]
Different types of data including mutation status and gene expression did not have a significant prognostic power.[32] No correlation to survival was observed in 72% of all studies with KRAS sequencing performed in non-small cell lung cancer (NSCLC).[32] However, KRAS mutations can not only affect the gene itself and the expression of the corresponding protein, but can also influence the expression of other downstream genes involved in crucial pathways regulating cell growth, differentiation and apoptosis. The different expression of these genes in KRAS-mutant tumors might have a more prominent role in affecting patient’s clinical outcomes.[32]
A 2008 paper published in Cancer Research concluded that the in vivo administration of the compound oncrasin-1 "suppressed the growth of K-ras mutant human lung tumor xenografts by >70% and prolonged the survival of nude mice bearing these tumors, without causing detectable toxicity", and that the "results indicate that oncrasin-1 or its active analogues could be a novel class of anticancer agents which effectively kill K-Ras mutant cancer cells."[33]
KRAS testing
In July 2009, the US Food and Drug Administration (FDA) updated the labels of two anti-EGFR monoclonal antibody drugs indicated for treatment of metastatic colorectal cancer, panitumumab (Vectibix) and cetuximab (Erbitux), to include information about KRAS mutations.[34]
In 2012, the FDA also cleared QIAGEN's therascreen KRAS test, which is a genetic test designed to detect the presence of seven mutations in the KRAS gene in colorectal cancer cells. This test is used to aid physicians in identifying patients with metastatic colorectal cancer for treatment with Erbitux. The presence of KRAS mutations in colorectal cancer tissue indicates that the patient may not benefit from treatment with Erbitux. If the test result indicates that the KRAS mutations are absent in the colorectal cancer cells, then the patient may be considered for treatment with Erbitux.[35]
As a drug target
Driver mutations in KRAS underlie the pathogenesis of up to 20% of human cancers.[36] Hence KRAS is an attractive drug target, however lack of obvious binding sites has hindered pharmaceutical development.[37] One potential drug interaction site is where GTP/GDP binds. However due to the extraordinarily high affinity of GTP/GDP for this site, it is unlikely that drug-like small molecule inhibitors could compete with GTP/GDP binding. Other than where GTP/GDP binds, there are no obvious high affinity binding sites for small molecules.[38]
G12C mutation
One fairly frequent driver mutation is KRASG12C which is adjacent a shallow binding site. This has allowed the development of electrophilic KRAS inhibitors that can form irreversible covalent bonds with nucleophilic sulfur atom of Cys-12 and hence selectively target KRASG12C and leave wild-type KRAS untouched.[39] Two KRASG12C mutant covalent inhibitors have reached clinical testing: AMG 510 (Amgen)[40] and MRTX-849 (Mirati Therapeutics)[41][42] while ARS-3248 (Wellspring Biosciences/Janssen) has received an investigational new drug (IND) approval to start clinical trials.[43] An antisense oligonucleotide (ASO), AZD4785 (AstraZeneca/Ionis Therapeutics) targeting KRAS has completed a phase I study[44] but was discontinued from further development because of insufficient knockdown of the target.[45]
Interactions
KRAS has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000133703 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000030265 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Tsuchida N, Ryder T, Ohtsubo E (1982). "Nucleotide sequence of the oncogene encoding p21 transforming protein of Kirsten murine sarcoma virus". Science. 217 (4563): 937–939. doi:10.1126/science.6287573. PMID 6287573.
- Scolnick EM, Papageoege AG, Shih TY (1979). "Guanine nucleotide-binding activity for src protein of rat-derived murine sarcoma viruses". Proc Natl Acad Sci USA. 76 (5): 5355–5559. doi:10.1073/pnas.76.10.5355. PMID 228228.
- Kranenburg O (November 2005). "The KRAS oncogene: past, present, and future". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1756 (2): 81–2. doi:10.1016/j.bbcan.2005.10.001. PMID 16269215.
- Welman A, Burger MM, Hagmann J (September 2000). "Structure and function of the C-terminal hypervariable region of K-Ras4B in plasma membrane targeting and transformation". Oncogene. 19 (40): 4582–91. doi:10.1038/sj.onc.1203818. PMID 11030147.
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N (September 2009). "Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells". Science. 325 (5947): 1555–9. doi:10.1126/science.1174229. PMC 2820374. PMID 19661383.
- Chiosea SI, Sherer CK, Jelic T, Dacic S (December 2011). "KRAS mutant allele-specific imbalance in lung adenocarcinoma". Modern Pathology. 24 (12): 1571–7. doi:10.1038/modpathol.2011.109. PMID 21743433.
- Hartman DJ, Davison JM, Foxwell TJ, Nikiforova MN, Chiosea SI (October 2012). "Mutant allele-specific imbalance modulates prognostic impact of KRAS mutations in colorectal adenocarcinoma and is associated with worse overall survival". International Journal of Cancer. 131 (8): 1810–7. doi:10.1002/ijc.27461. PMID 22290300.
- Krasinskas AM, Moser AJ, Saka B, Adsay NV, Chiosea SI (October 2013). "KRAS mutant allele-specific imbalance is associated with worse prognosis in pancreatic cancer and progression to undifferentiated carcinoma of the pancreas". Modern Pathology. 26 (10): 1346–54. doi:10.1038/modpathol.2013.71. PMC 4128625. PMID 23599154.
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP (March 2006). "Germline KRAS mutations cause Noonan syndrome". Nature Genetics. 38 (3): 331–6. doi:10.1038/ng1748. PMID 16474405.
- Niihori T, Aoki Y, Narumi Y, Neri G, Cavé H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y (March 2006). "Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome". Nature Genetics. 38 (3): 294–6. doi:10.1038/ng1749. PMID 16474404.
- Burmer GC, Loeb LA (April 1989). "Mutations in the KRAS2 oncogene during progressive stages of human colon carcinoma". Proceedings of the National Academy of Sciences of the United States of America. 86 (7): 2403–7. doi:10.1073/pnas.86.7.2403. PMC 286921. PMID 2648401.
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M (May 1988). "Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes". Cell. 53 (4): 549–54. doi:10.1016/0092-8674(88)90571-5. PMID 2453289.
- Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, Lam WK, Chiu SW, Girard L, Minna JD, Gazdar AF, Wong MP (March 2006). "Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features". Clinical Cancer Research. 12 (5): 1647–53. doi:10.1158/1078-0432.CCR-05-1981. PMID 16533793.
- Vogelstein B, Kinzler KW (August 2004). "Cancer genes and the pathways they control". Nature Medicine. 10 (8): 789–99. doi:10.1038/nm1087. PMID 15286780.
- Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S (June 2012). "Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum". Gut. 61 (6): 847–54. doi:10.1136/gutjnl-2011-300865. PMC 3345105. PMID 22427238.
- Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR, Jenkins MA, Win AK, Southey MC, Parry S, Hopper JL, Giles GG, Williamson E, English DR, Buchanan DD (June 2013). "Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features". Modern Pathology. 26 (6): 825–34. doi:10.1038/modpathol.2012.240. PMID 23348904.
- Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P (April 2006). "KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer". Cancer Research. 66 (8): 3992–5. doi:10.1158/0008-5472.CAN-06-0191. PMID 16618717.
- van Epps HL (Winter 2008). "Bittersweet Gene". CURE (Cancer Updates, Research and Education). Archived from the original on 2009-02-07.
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (February 2009). "Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer" (PDF). Journal of Clinical Oncology. 27 (5): 663–71. doi:10.1200/JCO.2008.20.8397. hdl:2434/652169. PMID 19114683.
- Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (April 2009). "Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer". The New England Journal of Medicine. 360 (14): 1408–17. doi:10.1056/NEJMoa0805019. PMID 19339720.
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M, Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C, Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D, Bardelli A (June 2012). "Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer". Nature. 486 (7404): 532–6. doi:10.1038/nature11156. PMC 3927413. PMID 22722830.
- Valtorta E, Misale S, Sartore-Bianchi A, Nagtegaal ID, Paraf F, Lauricella C, Dimartino V, Hobor S, Jacobs B, Ercolani C, Lamba S, Scala E, Veronese S, Laurent-Puig P, Siena S, Tejpar S, Mottolese M, Punt CJ, Gambacorta M, Bardelli A, Di Nicolantonio F (September 2013). "KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy". International Journal of Cancer. 133 (5): 1259–65. doi:10.1002/ijc.28106. PMID 23404247.
- Sankaranarayanan P, Schomay TE, Aiello KA, Alter O (April 2015). "Tensor GSVD of patient- and platform-matched tumor and normal DNA copy-number profiles uncovers chromosome arm-wide patterns of tumor-exclusive platform-consistent alterations encoding for cell transformation and predicting ovarian cancer survival". PLOS ONE. 10 (4): e0121396. doi:10.1371/journal.pone.0121396. PMC 4398562. PMID 25875127. AAAS EurekAlert! Press Release and NAE Podcast Feature.
- Chen Y, McGee J, Chen X, Doman TN, Gong X, Zhang Y, Hamm N, Ma X, Higgs RE, Bhagwat SV, Buchanan S, Peng SB, Staschke KA, Yadav V, Yue Y, Kouros-Mehr H (2014). "Identification of druggable cancer driver genes amplified across TCGA datasets". PLOS ONE. 9 (5): e98293. doi:10.1371/journal.pone.0098293. PMC 4038530. PMID 24874471.
- Suda K, Tomizawa K, Mitsudomi T (March 2010). "Biological and clinical significance of KRAS mutations in lung cancer: an oncogenic driver that contrasts with EGFR mutation". Cancer Metastasis Reviews. 29 (1): 49–60. doi:10.1007/s10555-010-9209-4. PMID 20108024.
- Riely GJ, Marks J, Pao W (April 2009). "KRAS mutations in non-small cell lung cancer". Proceedings of the American Thoracic Society. 6 (2): 201–5. doi:10.1513/pats.200809-107LC. PMID 19349489.
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE (January 2005). "KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib". PLOS Medicine. 2 (1): e17. doi:10.1371/journal.pmed.0020017. PMC 545207. PMID 15696205.
- Nagy Á, Pongor LS, Szabó A, Santarpia M, Győrffy B (February 2017). "KRAS driven expression signature has prognostic power superior to mutation status in non-small cell lung cancer". International Journal of Cancer. 140 (4): 930–937. doi:10.1002/ijc.30509. PMC 5299512. PMID 27859136.
- Guo W, Wu S, Liu J, Fang B (Sep 2008). "Identification of a small molecule with synthetic lethality for K-ras and protein kinase C iota". Cancer Research. 68 (18): 7403–8. doi:10.1158/0008-5472.CAN-08-1449. PMC 2678915. PMID 18794128.
- OncoGenetics.Org (July 2009). "FDA updates Vectibix and Erbitux labels with KRAS testing info". OncoGenetics.Org. Archived from the original on November 9, 2014. Retrieved 2009-07-20.
- FDA: Medical devices: therascreen® KRAS RGQ PCR Kit – P110030, accessed 20 Jone 2014
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ (November 2014). "Drugging the undruggable RAS: Mission possible?". Nature Reviews. Drug Discovery. 13 (11): 828–51. doi:10.1038/nrd4389. PMC 4355017. PMID 25323927.
- Ryan MB, Corcoran RB (November 2018). "Therapeutic strategies to target RAS-mutant cancers". Nature Reviews. Clinical Oncology. 15 (11): 709–720. doi:10.1038/s41571-018-0105-0. PMID 30275515.
- Holderfield M (July 2018). "Efforts to Develop KRAS Inhibitors". Cold Spring Harbor Perspectives in Medicine. 8 (7): a031864. doi:10.1101/cshperspect.a031864. PMID 29101115.
- McCormick F (January 2019). "Progress in targeting RAS with small molecule drugs". The Biochemical Journal. 476 (2): 365–374. doi:10.1042/BCJ20170441. PMID 30705085.
- Clinical trial number NCT03600883 for "A Phase 1/2, Study Evaluating the Safety, Tolerability, PK, and Efficacy of AMG 510 in Subjects With Solid Tumors With a Specific KRAS Mutation." at ClinicalTrials.gov
- Clinical trial number NCT03785249 for "MRTX849 in Patients With Cancer Having a KRAS G12C Mutation" at ClinicalTrials.gov
- Kaiser J (2019-10-30). "Two new drugs finally hit 'undruggable' cancer target, providing hope for treatments". Science Magazine. AAAS. Retrieved 2019-11-04.
- Mullard A (July 2019). "KRAS's undruggability cracks?". Nature Reviews. Drug Discovery. 18 (7): 488. doi:10.1038/d41573-019-00102-y. PMID 31267080.
- Clinical trial number NCT03101839 for "Phase I Dose-Escalation Study of AZD4785 in Patients With Advanced Solid Tumours" at ClinicalTrials.gov
- Plieth J (26 April 2019). "Astra's first attempt fails, but there's no giving up on KRAS". Evaluate.
- Li W, Han M, Guan KL (April 2000). "The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf". Genes & Development. 14 (8): 895–900. PMC 316541. PMID 10783161.
- Kiyono M, Kato J, Kataoka T, Kaziro Y, Satoh T (September 2000). "Stimulation of Ras guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) upon tyrosine phosphorylation by the Cdc42-regulated kinase ACK1". The Journal of Biological Chemistry. 275 (38): 29788–93. doi:10.1074/jbc.M001378200. PMID 10882715.
- Rubio I, Wittig U, Meyer C, Heinze R, Kadereit D, Waldmann H, Downward J, Wetzker R (November 1999). "Farnesylation of Ras is important for the interaction with phosphoinositide 3-kinase gamma". European Journal of Biochemistry. 266 (1): 70–82. doi:10.1046/j.1432-1327.1999.00815.x. PMID 10542052.
- Spaargaren M, Bischoff JR (December 1994). "Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap". Proceedings of the National Academy of Sciences of the United States of America. 91 (26): 12609–13. doi:10.1073/pnas.91.26.12609. PMC 45488. PMID 7809086.
- Vos MD, Ellis CA, Elam C, Ulku AS, Taylor BJ, Clark GJ (July 2003). "RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor". The Journal of Biological Chemistry. 278 (30): 28045–51. doi:10.1074/jbc.M300554200. PMID 12732644.
- Villalonga P, López-Alcalá C, Bosch M, Chiloeches A, Rocamora N, Gil J, Marais R, Marshall CJ, Bachs O, Agell N (November 2001). "Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling". Molecular and Cellular Biology. 21 (21): 7345–54. doi:10.1128/MCB.21.21.7345-7354.2001. PMC 99908. PMID 11585916.
Further reading
- Tsuchida N, Murugan AK, Grieco M (2016). "Kirsten Ras oncogene : significance of its discovery in human cancer research". Oncotarget. 7 (29): 46717–33. doi:10.18632/oncotarget.8773. PMC 5216832. PMID 27102293.
- Kahn S, Yamamoto F, Almoguera C, Winter E, Forrester K, Jordano J, Perucho M (1987). "The c-K-ras gene and human cancer (review)". Anticancer Research. 7 (4A): 639–52. PMID 3310850.
- Yamamoto F, Nakano H, Neville C, Perucho M (1985). "Structure and mechanisms of activation of c-K-ras oncogenes in human lung cancer". Progress in Medical Virology. Fortschritte der Medizinischen Virusforschung. Progres en Virologie Medicale. 32: 101–14. PMID 3895297.
- Porta M, Ayude D, Alguacil J, Jariod M (February 2003). "Exploring environmental causes of altered ras effects: fragmentation plus integration?". Molecular Carcinogenesis. 36 (2): 45–52. doi:10.1002/mc.10093. PMID 12557259.
- Smakman N, Borel Rinkes IH, Voest EE, Kranenburg O (November 2005). "Control of colorectal metastasis formation by K-Ras". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1756 (2): 103–14. doi:10.1016/j.bbcan.2005.07.001. PMID 16098678.
- Castagnola P, Giaretti W (November 2005). "Mutant KRAS, chromosomal instability and prognosis in colorectal cancer". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1756 (2): 115–25. doi:10.1016/j.bbcan.2005.06.003. PMID 16112461.
- Deramaudt T, Rustgi AK (November 2005). "Mutant KRAS in the initiation of pancreatic cancer". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1756 (2): 97–101. doi:10.1016/j.bbcan.2005.08.003. PMID 16169155.
- Pretlow TP, Pretlow TG (November 2005). "Mutant KRAS in aberrant crypt foci (ACF): initiation of colorectal cancer?". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1756 (2): 83–96. doi:10.1016/j.bbcan.2005.06.002. PMID 16219426.
- Su YH, Wang M, Aiamkitsumrit B, Brenner DE, Block TM (2005). "Detection of a K-ras mutation in urine of patients with colorectal cancer". Cancer Biomarkers. 1 (2–3): 177–82. doi:10.3233/CBM-2005-12-305. PMID 17192038.
- Domagała P, Hybiak J, Sulżyc-Bielicka V, Cybulski C, Ryś J, Domagała W (November 2012). "KRAS mutation testing in colorectal cancer as an example of the pathologist's role in personalized targeted therapy: a practical approach". Polish Journal of Pathology. 63 (3): 145–64. arXiv:1305.1286. doi:10.5114/PJP.2012.31499. PMID 23161231.
External links
- KRAS Reference Standards - Learn more about KRAS Reference Controls
- GeneReviews/NCBI/NIH/UW entry on Cardiofaciocutaneous Syndrome
- GeneReviews/NCBI/NIH/UW entry on Noonan syndrome
- KRAS2+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P01116 (Human GTPase KRas) at the PDBe-KB.