Geranyltranstransferase
In enzymology, a geranyltranstransferase (EC 2.5.1.10) is an enzyme that catalyzes the chemical reaction
- geranyl diphosphate + isopentenyl diphosphate diphosphate + trans,trans-farnesyl diphosphate
| geranyltranstransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.5.1.10 | ||||||||
| CAS number | 37277-79-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Thus, the two substrates of this enzyme are geranyl diphosphate (a 10 carbon precursor) and isopentenyl diphosphate (a 5 carbon precursor) whereas its two products are diphosphate and trans,trans-farnesyl diphosphate (a 15 carbon product).[1]
This enzyme belongs to the family of transferases, specifically those transferring aryl or alkyl groups other than methyl groups.
Nomenclature
The systematic name of this enzyme class is geranyl-diphosphate:isopentenyl-diphosphate geranyltranstransferase. Other names in common use include:
- farnesyl-diphosphate synthase
- geranyl transferase I
- prenyltransferase
- farnesyl pyrophosphate synthetase
- farnesylpyrophosphate synthetase
Common abbreviations include: FPS, FDS, FPPS, and FDPS.
Structure
The structure and mechanism of farnesyl pyrophosphate synthase (FPPS), a type of geranyltranstransferase, is well characterized. FPPS is a ~30 kDa Mg2+ dependent homodimeric enzyme that synthesizes (E, E)-farnesyl pyrophosphate in a successive manner from two equivalents of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP).[2]
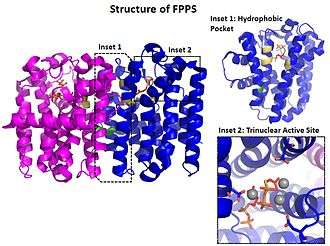
FPPS adopts a 3-layered α-helical fold characteristic of many prenyltransferases with 11 helices and flexible loops in between. The centrally located helices (α4 and α8) contain conserved aspartate motifs (DDXXD) that participate in substrate binding and catalysis.[3] Motif aspartate residues, water oxygens, and pyrophosphate coordinate three Mg2+in an octahedral manner. The trinuclear Mg2+ complex is critical for binding DMAPP and stabilizing the pyrophosphate leaving group while the growing hydrocarbon tail wedges into a deep hydrophobic pocket.[2] Site-directed mutagenesis studies have shown that the ultimate length of the isoprenoid product is determined by bulky residues (often phenyalanine) at the hydrophobic pocket's base.[4]
Mechanism
From crystal structures and kinetic assays, it is believed that FPPS catalyzes the condensation reaction in three concerted steps: (1) Ionization, (2) Condensation, and (3) Elimination.[2]
In the first step, three Mg2+stabilize the anionic leaving group, pyrophosphate, on dimethylallyl pyrophosphate (DMAPP). The loss of pyrophosphate forms an allylic carbocation on dimethylallyl. In the second step, the reactive C3-C5 double bond in isopentyl pyrophosphate (IPP) nucleophilically attacks the previously formed dimethylallyl carbocation in a 5-carbon/5-carbon condensation reaction. The final step involves pyrophosphate held in the trinuclear Mg2+ center acting as a catalytic base in an elimination reaction to form geranyl pyrophosphate. A second consecutive round of geranyl pyrophosphate ionization, condensation with IPP, and elimination forms farnesyl pyrophosphate.[2][5]
Function
Geranyltranstransferases are an evolutionarily conserved class of enzymes in Archaea, Bacteria, and Eukarya that participate in a broad range of biosynthetic pathways including those of cholesterol, porphyrin, carotenoids, ubiquinone, and isoprenoids.[3] Various studies have located FPPS in chloroplasts, mitochondria, cytosol, and peroxisomes.[6][7][8]
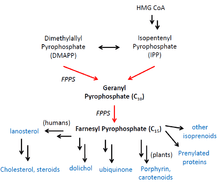
In cholesterol synthesis, the product, farnesyl pyrophosphate, is consumed in a reductive tail-to-tail condensation with another farnesyl pyrophosphate to form a 30-carbon compound called squalene by squalene synthase.[9] Through several more biosynthetic steps, squalene is transformed into lanosterol, a direct precursor for cholesterol.[10] Notably, sterols control FPPS expression through two cis regulatory factors (an inverted CAAT box and SRE-3) in the proximal FPPS promoter.[11] In plants, porphyrin and carotenoids constitute accessory pigments that help capture light in the photosystems. Ubiquinone is a key electron carrier in the electron transport chain of cellular respiration. Isoprenoids are a large group of compounds that serve as biosynthetic precursors for lipids and hormones.[6]
Farnesyl and geranyl pyrophosphate also serve as precursors for prenylated proteins. Prenylation is a common type of covalent post-translational modification at C-terminal CaaX motifs that allows proteins to localize to membranes or bind to one another. A notable example of the former is the farnesylation of small G-proteins including Ras, CDC42, Rho, and Rac. The attachment of a hydrophobic aliphatic chain as those present in farnesyl or geranylgeranyl groups allows small G-proteins to tether from membranes and carry out effector functions.[12]
Drug targeting
FPPS is the target of bisphosphonate drugs such as Fosamax (alendronate) and Actonel (risedronate). Bisphosphonate drugs are commonly prescribed for bone diseases including Paget’s disease, osteolytic metastases, and post-menopausal osteoporosis. Bisphosphonate drugs help maintain bone tissue in osteoporotic patients and reduce blood calcium levels in hypercalcemic patients by inhibiting FPPS in bone-reabsorbing osteoclasts. An FPPS-IPP-risendronate ternary complex demonstrated that risendronate binds to the trinuclear Mg2+ complex and interacts with the hydrophobic pocket in a manner similar to DMAPP.[2][13]
References
- Lynen F, Agranoff BW, Eggerer H, Henning U, Möslein EM (1959). "Zur Biosynthese der Terpene. VI gamma,gamma-Dimethyl-allyl-pyrophosphat und Geranyl-pyrophosphat, biologische Vorstufen des Squalens". Angew. Chem. 71 (21): 657–663. doi:10.1002/ange.19590712102.
- Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J (Mar 2004). "Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis". The Journal of Biological Chemistry. 279 (10): 8526–9. doi:10.1074/jbc.C300511200. PMID 14672944.
- Chen A, Kroon PA, Poulter CD (Apr 1994). "Isoprenyl diphosphate synthases: protein sequence comparisons, a phylogenetic tree, and predictions of secondary structure". Protein Science. 3 (4): 600–7. doi:10.1002/pro.5560030408. PMC 2142870. PMID 8003978.
- Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD (Dec 1996). "Regulation of product chain length by isoprenyl diphosphate synthases". Proceedings of the National Academy of Sciences of the United States of America. 93 (26): 15018–23. doi:10.1073/pnas.93.26.15018. PMC 26348. PMID 8986756.
- Tarshis LC, Yan M, Poulter CD, Sacchettini JC (Sep 1994). "Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-A resolution". Biochemistry. 33 (36): 10871–7. doi:10.1021/bi00202a004. PMID 8086404.
- Dhar MK, Koul A, Kaul S (Jan 2013). "Farnesyl pyrophosphate synthase: a key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development". New Biotechnology. 30 (2): 114–23. doi:10.1016/j.nbt.2012.07.001. PMID 22842101.
- Sanmiya K, Ueno O, Matsuoka M, Yamamoto N (Mar 1999). "Localization of farnesyl diphosphate synthase in chloroplasts". Plant & Cell Physiology. 40 (3): 348–54. doi:10.1093/oxfordjournals.pcp.a029549. PMID 10353221.
- Martín D, Piulachs MD, Cunillera N, Ferrer A, Bellés X (Mar 2007). "Mitochondrial targeting of farnesyl diphosphate synthase is a widespread phenomenon in eukaryotes". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1773 (3): 419–26. doi:10.1016/j.bbamcr.2006.11.015. PMID 17198737.
- Tansey TR, Shechter I (2001). "Squalene synthase: structure and regulation". Prog. Nucleic Acid Res. Mol. Biol. Progress in Nucleic Acid Research and Molecular Biology. 65: 157–95. doi:10.1016/S0079-6603(00)65005-5. ISBN 9780125400657. PMID 11008488.
- Liang PH, Ko TP, Wang AH (2002). "Structure, mechanism and function of prenyltransferases". Eur. J. Biochem. 269 (14): 3339–54. doi:10.1046/j.1432-1033.2002.03014.x. PMID 12135472.
- Ericsson J, Jackson SM, Edwards PA (Oct 1996). "Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene". The Journal of Biological Chemistry. 271 (40): 24359–64. doi:10.1074/jbc.271.40.24359. PMID 8798690.
- Sebti SM (Apr 2005). "Protein farnesylation: implications for normal physiology, malignant transformation, and cancer therapy". Cancer Cell. 7 (4): 297–300. doi:10.1016/j.ccr.2005.04.005. PMID 15837619.
- Guo RT, Cao R, Liang PH, Ko TP, Chang TH, Hudock MP, Jeng WY, Chen CK, Zhang Y, Song Y, Kuo CJ, Yin F, Oldfield E, Wang AH (Jun 2007). "Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases". Proceedings of the National Academy of Sciences of the United States of America. 104 (24): 10022–7. doi:10.1073/pnas.0702254104. PMC 1877987. PMID 17535895.