BRCA2
BRCA2 and BRCA2 (/ˌbrækəˈtuː/[5]) are a human gene and its protein product, respectively. The official symbol (BRCA2, italic for the gene, nonitalic for the protein) and the official name (originally breast cancer 2; currently BRCA2, DNA repair associated) are maintained by the HUGO Gene Nomenclature Committee. One alternative symbol, FANCD1, recognizes its association with the FANC protein complex. Orthologs, styled Brca2 and Brca2, are common in other vertebrate species.[6][7] BRCA2 is a human tumor suppressor gene[8][9] (specifically, a caretaker gene), found in all humans; its protein, also called by the synonym breast cancer type 2 susceptibility protein, is responsible for repairing DNA.[10]
| BRCA2 repeat | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of a rad51-brca2 brc repeat complex | |||||||||
| Identifiers | |||||||||
| Symbol | BRCA2 | ||||||||
| Pfam | PF00634 | ||||||||
| InterPro | IPR002093 | ||||||||
| SCOPe | 1n0w / SUPFAM | ||||||||
| |||||||||
| BRCA-2 helical | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of a brca2-dss1 complex | |||||||||
| Identifiers | |||||||||
| Symbol | BRCA-2_helical | ||||||||
| Pfam | PF09169 | ||||||||
| InterPro | IPR015252 | ||||||||
| SCOPe | 1iyj / SUPFAM | ||||||||
| |||||||||
| BRCA2, oligonucleotide/oligosaccharide-binding, domain 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of a brca2-dss1 complex | |||||||||
| Identifiers | |||||||||
| Symbol | BRCA-2_OB1 | ||||||||
| Pfam | PF09103 | ||||||||
| InterPro | IPR015187 | ||||||||
| SCOPe | 1iyj / SUPFAM | ||||||||
| |||||||||
| BRCA2, oligonucleotide/oligosaccharide-binding, domain 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of a brca2-dss1 complex | |||||||||
| Identifiers | |||||||||
| Symbol | BRCA-2_OB3 | ||||||||
| Pfam | PF09104 | ||||||||
| InterPro | IPR015188 | ||||||||
| SCOPe | 1iyj / SUPFAM | ||||||||
| |||||||||
| Tower domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 structure of a brca2-dss1 complex | |||||||||
| Identifiers | |||||||||
| Symbol | Tower | ||||||||
| Pfam | PF09121 | ||||||||
| InterPro | IPR015205 | ||||||||
| SCOPe | 1mje / SUPFAM | ||||||||
| |||||||||
BRCA2 and BRCA1 are normally expressed in the cells of breast and other tissue, where they help repair damaged DNA or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double strand breaks.[11][12] If BRCA1 or BRCA2 itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer.[13][14] BRCA1 and BRCA2 have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a normal tumor suppressive function whereas high penetrance mutations in these genes cause a loss of tumor suppressive function, which correlates with an increased risk of breast cancer.[15]
The BRCA2 gene is located on the long (q) arm of chromosome 13 at position 12.3 (13q12.3).[16] The human reference BRCA 2 gene contains 27 exons, and the cDNA has 10,254 base pairs[17] coding for a protein of 3418 amino acids.[18][19]
Function
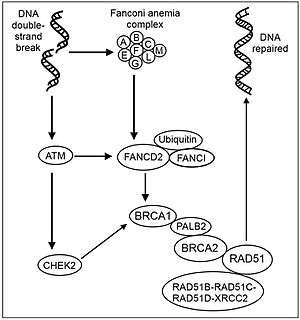
Although the structures of the BRCA1 and BRCA2 genes are very different, at least some functions are interrelated. The proteins made by both genes are essential for repairing damaged DNA (see Figure of recombinational repair steps). BRCA2 binds the single strand DNA and directly interacts with the recombinase RAD51 to stimulate[27] and maintain [28] strand invasion, a vital step of homologous recombination. The localization of RAD51 to the DNA double-strand break requires the formation of the BRCA1-PALB2-BRCA2 complex. PALB2 (Partner and localizer of BRCA2)[29] can function synergistically with a BRCA2 chimera (termed piccolo, or piBRCA2) to further promote strand invasion.[30] These breaks can be caused by natural and medical radiation or other environmental exposures, but also occur when chromosomes exchange genetic material during a special type of cell division that creates sperm and eggs (meiosis). Double strand breaks are also generated during repair of DNA cross links. By repairing DNA, these proteins play a role in maintaining the stability of the human genome and prevent dangerous gene rearrangements that can lead to hematologic and other cancers.
BRCA2 has been shown to possess a crucial role in protection from the MRE11-dependent nucleolytic degradation of the reversed forks that are forming during DNA replication fork stalling (caused by obstacles such as mutations, intercalating agents etc.).[31]
Like BRCA1, BRCA2 probably regulates the activity of other genes and plays a critical role in embryo development.
Clinical significance
Certain variations of the BRCA2 gene increase risks for breast cancer as part of a hereditary breast–ovarian cancer syndrome. Researchers have identified hundreds of mutations in the BRCA2 gene, many of which cause an increased risk of cancer. BRCA2 mutations are usually insertions or deletions of a small number of DNA base pairs in the gene. As a result of these mutations, the protein product of the BRCA2 gene is abnormal, and does not function properly. Researchers believe that the defective BRCA2 protein is unable to fix DNA damage that occurs throughout the genome. As a result, there is an increase in mutations due to error-prone translesion synthesis past un-repaired DNA damage, and some of these mutations can cause cells to divide in an uncontrolled way and form a tumor.
People who have two mutated copies of the BRCA2 gene have one type of Fanconi anemia. This condition is caused by extremely reduced levels of the BRCA2 protein in cells, which allows the accumulation of damaged DNA. Patients with Fanconi anemia are prone to several types of leukemia (a type of blood cell cancer); solid tumors, particularly of the head, neck, skin, and reproductive organs; and bone marrow suppression (reduced blood cell production that leads to anemia). Women having inherited a defective BRCA1 or BRCA2 gene have risks for breast and ovarian cancer that are so high and seem so selective that many mutation carriers choose to have prophylactic surgery. There has been much conjecture to explain such apparently striking tissue specificity. Major determinants of where BRCA1- and BRCA2-associated hereditary cancers occur are related to tissue specificity of the cancer pathogen, the agent that causes chronic inflammation, or the carcinogen. The target tissue may have receptors for the pathogen, become selectively exposed to carcinogens and an infectious process. An innate genomic deficit impairs normal responses and exacerbates the susceptibility to disease in organ targets. This theory also fits data for several tumor suppressors beyond BRCA1 or BRCA2. A major advantage of this model is that it suggests there are some options in addition to prophylactic surgery.[32]
In addition to breast cancer in men and women, mutations in BRCA2 also lead to an increased risk of ovarian, Fallopian tube, prostate and pancreatic cancer. In some studies, mutations in the central part of the gene have been associated with a higher risk of ovarian cancer and a lower risk of prostate cancer than mutations in other parts of the gene. Several other types of cancer have also been seen in certain families with BRCA2 mutations.
In general, strongly inherited gene mutations (including mutations in BRCA2) account for only 5-10% of breast cancer cases; the specific risk of getting breast or other cancer for anyone carrying a BRCA2 mutation depends on many factors.[33]
History
| The BRCA2 gene was discovered in 1994.[34][16][35]
The gene was first cloned by scientists at Myriad Genetics, Endo Recherche, Inc., HSC Research & Development Limited Partnership, and the University of Pennsylvania.[36] Methods to diagnose the likelihood of a patient with mutations in BRCA1 and BRCA2 getting cancer were covered by patents owned or controlled by Myriad Genetics.[37][38] Myriad's business model of exclusively offering the diagnostic test led from Myriad's beginnings as a startup in 1994 to its being a publicly traded company with 1200 employees and about $500M in annual revenue in 2012;[39] it also led to controversy over high test prices and the unavailability of second opinions from other diagnostic labs, which in turn led to the landmark Association for Molecular Pathology v. Myriad Genetics lawsuit.[40] Germline BRCA2 mutations and founder effectAll germline BRCA2 mutations identified to date have been inherited, suggesting the possibility of a large "founder" effect in which a certain mutation is common to a well-defined population group and can theoretically be traced back to a common ancestor. Given the complexity of mutation screening for BRCA2, these common mutations may simplify the methods required for mutation screening in certain populations. Analysis of mutations that occur with high frequency also permits the study of their clinical expression.[41] A striking example of a founder mutation is found in Iceland, where a single BRCA2 (999del5) mutation accounts for virtually all breast/ovarian cancer families.[42][43] This frame-shift mutation leads to a highly truncated protein product. In a large study examining hundreds of cancer and control individuals, this 999del5 mutation was found in 0.6% of the general population. Of note, while 72% of patients who were found to be carriers had a moderate or strong family history of breast cancer, 28% had little or no family history of the disease. This strongly suggests the presence of modifying genes that affect the phenotypic expression of this mutation, or possibly the interaction of the BRCA2 mutation with environmental factors. Additional examples of founder mutations in BRCA2 are given in the table below.
MeiosisIn the plant Arabidopsis thaliana, loss of the BRCA2 homolog AtBRCA2 causes severe defects in both male meiosis and in the development of the female gametocyte.[59] AtBRCA2 protein is required for proper localization of the synaptonemal complex protein AtZYP1 and the recombinases AtRAD51 and AtDMC1. Furthermore, AtBRCA2 is required for proper meiotic synapsis. Thus AtBRCA2 is likely important for meiotic recombination. It appears that AtBRCA2 acts during meiosis to control the single-strand invasion steps mediated by AtRAD51 and AtDMC1 occurring during meiotic homologous recombinational repair of DNA damages.[59] Homologs of BRCA2 are also essential for meiosis in the fungus Ustilago maydis,[60] the worm Caenorhabditis elegans,[61][62] and the fruitfly Drosophila melanogaster.[63] Mice that produce truncated versions of BRCA2 are viable but sterile.[64] BRCA2 mutant rats have a phenotype of growth inhibition and sterility in both sexes.[65] Aspermatogenesis in these mutant rats is due to a failure of homologous chromosome synapsis during meiosis. BRC repeat sequencesDMC1 (DNA meiotic recombinase 1) is a meiosis specific homolog of RAD51 that mediates strand exchange during homologous recombinational repair. DMC1 promotes the formation of DNA strand invasion products (joint molecules) between homologous DNA molecules. Human DMC1 interacts directly with each of a series of repeat sequences in the BRCA2 protein (called BRC repeats) that stimulate joint molecule formation by DMC1.[66] BRC repeats conform to a motif consisting of a sequence of about 35 highly conserved amino acids that are present at least once in all BRCA2-like proteins. The BRCA2 BRC repeats stimulate joint molecule formation by promoting the interaction of single-stranded DNA (ssDNA) with DMC1.[66] The ssDNA complexed with DMC1 can pair with homologous ssDNA from another chromosome during the synopsis stage of meiosis to form a joint molecule, a central step in homologous recombination. Thus the BRC repeat sequences of BRCA2 appear to play a key role in recombinational repair of DNA damages during meiotic recombination. Overall, it appears that homologous recombination during meiosis functions to repair DNA damages, and that BRCA2 plays a key role in performing this function. NeurogenesisBRCA2 is required in the mouse for neurogenesis and suppression of medulloblastoma.[67] ‘’BRCA2’’ loss profoundly affects neurogenesis, particularly during embryonic and postnatal neural development. These neurological defects arise from DNA damage.[67] Epigenetic controlEpigenetic alterations in expression of BRCA2 (causing over-expression or under-expression) are very frequent in sporadic cancers (see Table below) while mutations in BRCA2 are rarely found.[68][69][70] In non-small cell lung cancer, BRCA2 is epigenetically repressed by hypermethylation of the promoter.[71] In this case, promoter hypermethylation is significantly associated with low mRNA expression and low protein expression but not with loss of heterozygosity of the gene. In sporadic ovarian cancer, an opposite effect is found. BRCA2 promoter and 5'-UTR regions have relatively few or no methylated CpG dinucleotides in the tumor DNA compared with that of non-tumor DNA, and a significant correlation is found between hypomethylation and a >3-fold over-expression of BRCA2.[72] This indicates that hypomethylation of the BRCA2 promoter and 5'-UTR regions leads to over-expression of BRCA2 mRNA. One report indicated some epigenetic control of BRCA2 expression by the microRNAs miR-146a and miR-148a.[73] BRCA2 expression in cancerIn eukaryotes, BRCA2 protein has an important role in homologous recombinational repair. In mice and humans, BRCA2 primarily mediates orderly assembly of RAD51 on single-stranded (ss) DNA, the form that is active for homologous pairing and strand invasion.[74] BRCA2 also redirects RAD51 from double-stranded DNA and prevents dissociation from ssDNA.[74] In addition, the four paralogs of RAD51, consisting of RAD51B (RAD51L1), RAD51C (RAD51L2), RAD51D (RAD51L3), XRCC2 form a complex called the BCDX2 complex (see Figure: Recombinational repair of DNA). This complex participates in RAD51 recruitment or stabilization at damage sites.[26] The BCDX2 complex appears to act by facilitating the assembly or stability of the RAD51 nucleoprotein filament. RAD51 catalyses strand transfer between a broken sequence and its undamaged homologue to allow re-synthesis of the damaged region (see homologous recombination models). Some studies of cancers report over-expressed BRCA2 whereas other studies report under-expression of BRCA2. At least two reports found over-expression in some sporadic breast tumors and under-expression in other sporadic breast tumors.[75][76] (see Table). Many cancers have epigenetic deficiencies in various DNA repair genes (see Frequencies of epimutations in DNA repair genes in cancers). These repair deficiencies likely cause increased unrepaired DNA damages. The over-expression of BRCA2 seen in many cancers may reflect compensatory BRCA2 over-expression and increased homologous recombinational repair to at least partially deal with such excess DNA damages. Egawa et al.[77] suggest that increased expression of BRCA2 can be explained by the genomic instability frequently seen in cancers, which induces BRCA2 mRNA expression due to an increased need for BRCA2 for DNA repair. Under-expression of BRCA2 would itself lead to increased unrepaired DNA damages. Replication errors past these damages (see translesion synthesis) would lead to increased mutations and cancer.
InteractionsBRCA2 has been shown to interact with
Domain architectureBRCA2 contains a number of 39 amino acid repeats that are critical for binding to RAD51 (a key protein in DNA recombinational repair) and resistance to methyl methanesulphonate treatment.[96][103][104][112] The BRCA2 helical domain adopts a helical structure, consisting of a four-helix cluster core (alpha 1, alpha 8, alpha 9, alpha 10) and two successive beta-hairpins (beta 1 to beta 4). An approximately 50-amino acid segment that contains four short helices (alpha 2 to alpha 4), meanders around the surface of the core structure. In BRCA2, the alpha 9 and alpha 10 helices pack with the BRCA2 OB1 domain through van der Waals contacts involving hydrophobic and aromatic residues, and also through side-chain and backbone hydrogen bonds. This domain binds the 70-amino acid DSS1 (deleted in split-hand/split foot syndrome) protein, which was originally identified as one of three genes that map to a 1.5-Mb locus deleted in an inherited developmental malformation syndrome.[110] The BRCA OB1 domain assumes an OB fold, which consists of a highly curved five-stranded beta-sheet that closes on itself to form a beta-barrel. OB1 has a shallow groove formed by one face of the curved sheet and is demarcated by two loops, one between beta 1 and beta 2 and another between beta 4 and beta 5, which allows for weak single strand DNA binding. The domain also binds the 70-amino acid DSS1 (deleted in split-hand/split foot syndrome) protein.[110] The BRCA OB3 domain assumes an OB fold, which consists of a highly curved five-stranded beta-sheet that closes on itself to form a beta-barrel. OB3 has a pronounced groove formed by one face of the curved sheet and is demarcated by two loops, one between beta 1 and beta 2 and another between beta 4 and beta 5, which allows for strong ssDNA binding.[110] The Tower domain adopts a secondary structure consisting of a pair of long, antiparallel alpha-helices (the stem) that support a three-helix bundle (3HB) at their end. The 3HB contains a helix-turn-helix motif and is similar to the DNA binding domains of the bacterial site-specific recombinases, and of eukaryotic Myb and homeodomain transcription factors. The Tower domain has an important role in the tumour suppressor function of BRCA2, and is essential for appropriate binding of BRCA2 to DNA.[110] Patents, enforcement, litigation, and controversyA patent application for the isolated BRCA1 gene and cancer-cancer promoting mutations, as well as methods to diagnose the likelihood of getting breast cancer, was filed by the University of Utah, National Institute of Environmental Health Sciences (NIEHS) and Myriad Genetics in 1994;[37] over the next year, Myriad, in collaboration with other investigators, isolated and sequenced the BRCA2 gene and identified relevant mutations, and the first BRCA2 patent was filed in the U.S. by Myriad and the other institutions in 1995.[36] Myriad is the exclusive licensee of these patents and has enforced them in the US against clinical diagnostic labs.[40] This business model led from Myriad being a startup in 1994 to being a publicly traded company with 1200 employees and about $500M in annual revenue in 2012;[39] it also led to controversy over high prices and the inability to get second opinions from other diagnostic labs, which in turn led to the landmark Association for Molecular Pathology v. Myriad Genetics lawsuit.[40][113] The patents begin to expire in 2014. Peter Meldrum, CEO of Myriad Genetics, has acknowledged that Myriad has "other competitive advantages that may make such [patent] enforcement unnecessary" in Europe.[114] Legal decisions surrounding the BRCA1 and BRCA2 patents will affect the field of genetic testing in general.[115] In June 2013, in Association for Molecular Pathology v. Myriad Genetics (No. 12-398), the US Supreme Court unanimously ruled that, "A naturally occurring DNA segment is a product of nature and not patent eligible merely because it has been isolated," invalidating Myriad's patents on the BRCA1 and BRCA2 genes. However, the Court also held that manipulation of a gene to create something not found in nature could still be eligible for patent protection.[116] The Federal Court of Australia came to the opposite conclusion, upholding the validity of an Australian Myriad Genetics patent over the BRCA1 gene in February 2013,[117] but this decision is being appealed and the appeal will include consideration of the US Supreme Court ruling.[118] References
Further reading
External links
|