Synaptonemal complex
The synaptonemal complex (SC) is a protein structure that forms between homologous chromosomes (two pairs of sister chromatids) during meiosis and is thought to mediate synapsis and recombination during meiosis I in eukaryotes. It is currently thought that the SC functions primarily as a scaffold to allow interacting chromatids to complete their crossover activities[1].
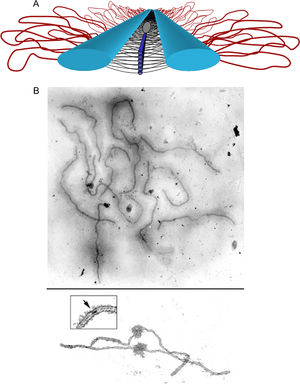
Composition
The synaptonemal complex is a tripartite structure consisting of two parallel lateral regions and a central element. This "tripartite structure" is seen during the pachytene stage of the first meiotic prophase, both in males and in females during gametogenesis. Previous to the pachytene stage, during leptonema, the lateral elements begin to form and they initiate and complete their pairing during the zygotene stage. After pachynema ends, the SC usually becomes disassembled and can no longer be identified[2].
In humans, three specific components of the synaptonemal complex have been characterized: SC protein-1 (SYCP1), SC protein-2 (SYCP2), and SC protein-3 (SYCP3). The SYCP1 gene is on chromosome 1p13; the SYCP2 gene is on chromosome 20q13.33; and the gene for SYCP3 is on chromosome 12q.[3]
The synaptonemal complex was described by Montrose J. Moses in 1956 in primary spermatocytes of crayfish and by D. Fawcett in spermatocytes of pigeon, cat and man[4]. As seen with the electron microscope, the synaptonemal complex is formed by two "lateral elements", mainly formed by SYCP3 and secondarily by SYCP2, a "central element" that contains at least two additional proteins and the amino terminal region of SYCP1, and a "central region" spanned between the two lateral elements, that contains the "transverse filaments" composed mainly by the protein SYCP1[3].
The SCs can be seen with the light microscope using silver staining or with immunofluorescence techniques that label the proteins SYCP3 or SYCP2.
Assembly and Disassembly
Formation of the SC usually reflects the pairing or "synapsis" of homologous chromosomes and may be used to probe the presence of pairing abnormalities in individuals carrying chromosomal abnormalities, either in number or in the chromosomal structure[5]. The sex chromosomes in male mammals show only "partial synapsis" as they usually form only a short SC in the XY pair. The SC shows very little structural variability among eukaryotic organisms despite some significant protein differences. In many organisms the SC carries one or several "recombination nodules" associated to its central space. These nodules are thought to correspond to mature genetic recombination events or "crossovers". In male mice, gamma irradiation increases meiotic crossovers in SCs. This indicates that exogenously caused DNA damages are likely repaired by crossover recombination in SCs[6]. The finding of an interaction between a SC structural component [synaptonemal central element protein 2 (SYCE2)] and recombinational repair protein RAD51 also suggests a role of the SC in DNA repair.
In cell development the synaptonemal complex disappears during the late prophase of meiosis I.It is formed during zygotene
Necessity in Eukaryotes
It is now evident that the synaptonemal complex is not required for genetic recombination in some organisms. For instance, in protozoan ciliates such as Tetrahymena thermophila and Paramecium tetraurelia genetic crossover does not appear to require synaptonemal complex formation[7][8]. Research has shown that not only does the SC form after genetic recombination but mutant yeast cells unable to assemble a synaptonemal complex can still engage in the exchange of genetic information. However, in other organisms like the C. elegans nematode, formation of chiasmata require the formation of the synaptonemal complex.
External links
| Wikimedia Commons has media related to Synaptonemal complex. |
by 3D-Structured Illumination, photograph by Dr. Chung-Ju Rachel Wang University of California Berkeley, Department of Molecular and Cell Biology Berkeley, CA, USA, second place winner of the 2009 Olympus Bioscapes Digital Imaging Competition.
References
- Page SL, Hawley RS (2004-10-08). "The genetics and molecular biology of the synaptonemal complex". Annual Review of Cell and Developmental Biology. 20 (1): 525–58. doi:10.1146/annurev.cellbio.19.111301.155141. PMID 15473851.
- Yang F, Wang PJ (2009). "The Mammalian synaptonemal complex: a scaffold and beyond". Genome Dynamics. 5: 69–80. doi:10.1159/000166620. ISBN 978-3-8055-8967-3. PMID 18948708.
- Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. (February 2009). "Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair". PLOS Genetics. 5 (2): e1000393. doi:10.1371/journal.pgen.1000393. PMC 2640461. PMID 19247432.
- Moses, Montrose J. (1968-12-01). "Synaptinemal complex". Annual Review of Genetics. 2 (1): 363–412. doi:10.1146/annurev.ge.02.120168.002051. ISSN 0066-4197.
- Zickler D, Kleckner N (1999-12-01). "Meiotic chromosomes: integrating structure and function". Annual Review of Genetics. 33 (1): 603–754. doi:10.1146/annurev.genet.33.1.603. PMID 10690419.
- Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. (February 2009). "Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair". PLOS Genetics. 5 (2): e1000393. doi:10.1371/journal.pgen.1000393. PMC 2640461. PMID 19247432.
- Lukaszewicz A, Howard-Till RA, Loidl J (November 2013). "Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex". Nucleic Acids Research. 41 (20): 9296–309. doi:10.1093/nar/gkt703. PMC 3814389. PMID 23935123.
- Chi J, Mahé F, Loidl J, Logsdon J, Dunthorn M (March 2014). "Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway". Molecular Biology and Evolution. 31 (3): 660–72. doi:10.1093/molbev/mst258. PMID 24336924.