QPNC-PAGE
QPNC-PAGE, or quantitative preparative native continuous polyacrylamide gel electrophoresis, is a bioanalytical, high-resolution and highly accurate technique applied in biochemistry and bioinorganic chemistry to separate proteins quantitatively by isoelectric point. This standardized variant of native gel electrophoresis is used by biologists to isolate biomacromolecules in solution, for example, active or native metalloproteins in biological samples or properly and improperly folded metal cofactor-containing proteins or protein isoforms in complex protein mixtures.[1]
Introduction
Proteins perform several functions in living organisms, including catalytic reactions and transport of molecules or ions within the cells, the organs or the whole body. The understanding of the processes in human organisms, which are mainly driven by biochemical reactions and protein-protein interactions, depends to a great extent on our ability to isolate active proteins in biological samples for more detailed examination of chemical structure and physiological function. This essential information can imply an important indication of a patient's state of health.[2]
As about 30-40% of all known proteins contain one or more metal ion cofactors (e.g., ceruloplasmin, ferritin, amyloid precursor protein, matrix metalloproteinase), especially native metalloproteins have to be isolated, identified and quantified after liquid biopsy. Many of these cofactors (e.g., iron, copper, or zinc) play a key role in vital enzymatic catalytic processes or stabilize globular protein molecules.[3] Therefore, the high-precision electrophoresis and other native separation techniques are highly relevant as initial step of protein and trace metal speciation analysis, subsequently, followed by mass spectrometric and magnetic resonance methods for quantifying and identifying the soluble proteins of interest.[4]
Method
Separation and buffering mechanisms

In gel electrophoresis proteins are normally separated by charge, size, or shape. The aim of isoelectric focusing (IEF), for example, is to separate proteins according to their isoelectric point (pI), thus, according to their charge at different pH values.[5] Here, the same mechanism is accomplished in a commercially available electrophoresis chamber (see figure Equipment) for separating charged biomolecules, for example, superoxide dismutase (SOD)[6] or allergens,[7] at continuous pH conditions and different velocities of migration depending on different isoelectric points. The separated (metal) proteins elute sequentially, starting with the lowest (pI > 2-4) and ending with the highest pI (pI < 10.0) of the present protein molecules.
Due to the specific properties of the prepared gel and electrophoresis buffer solution (which is basic and contains Tris-HCl and NaN3), most proteins of a biological system (e.g., Helicobacter pylori[8]) are charged negatively in the solution, and will migrate from the cathode to the anode due to the electric field. At the anode, electrochemically-generated hydrogen ions react with Tris molecules to form monovalent Tris ions. The positively charged Tris ions migrate through the gel to the cathode where they neutralise hydroxide ions to form Tris molecules and water. Thus, the Tris-based buffering mechanism causes a constant pH in the buffer system.[9] At 25 °C Tris buffer has an effective pH range between 7.5 and 9.0. Under the conditions given here (addressing the concentration, buffering mechanism, pH and temperature) the effective pH is shifted in the range of about 10.0 to 10.5. Native buffer systems all have low conductivity and range in pH from 3.8 to 10.2. Continuous native buffer systems are used to separate proteins according their pI.[10]
Although the pH value (10.00) of the electrophoresis buffer does not correspond to a physiological pH value within a cell or tissue type, the separated ring-shaped protein bands are eluted continuously into a physiological buffer solution (pH 8.00) and isolated in different fractions (see figure Electropherogram).[11] Provided that irreversible denaturation cannot be demonstrated (by an independent procedure), most protein molecules are stable in aqueous solution, at pH values from 3 to 10 if the temperature is below 50 °C.[12] As the Joule heat and temperature generated during electrophoresis may exceed 50 °C,[13] and thus, have a negative impact on the stability and migration behavior of proteins, the separation system, including the electrophoresis chamber and a fraction collector, is cooled in a refrigerator at 4 °C. Overheating of the gel is impeded by internal cooling of the gel column and by generating a constant power (see figure Equipment).
Gel properties and polymerization time
Best polymerization conditions for acrylamide gels are obtained at 25-30 °C[14] and polymerization seems terminated after 20-30 min of reaction although residual monomers (10-30%) are detected after this time.[15] The co-polymerization of acrylamide (AA) monomer/N,N'-Methylenebisacrylamide (Bis-AA) cross-linker initiated by ammonium persulfate (APS)/tetramethylethylenediamine (TEMED) reactions, is most efficient at alkaline pH. Thereby, acrylamide chains are created and cross-linked at a time. Due to the properties of the electrophoresis buffer the gel polymerization is conducted at pH 10.00 making sure an efficient use of TEMED and APS as catalysts of the polymerization reaction, and concurrently, suppressing a competitive hydrolysis of the produced acrylamide polymer network. Otherwise, proteins could be modified by reaction with unpolymerized monomers of acrylamide, forming covalent acrylamide adduction products that may result in multiple bands.[16]
Additionally, the time of polymerization of a gel may directly affect the peak-elution times of separated metalloproteins in the electropherogram due to the compression and dilatation of the gels and their pores with the longer incubation times (see figure Electropherogram, cf. Reproducibility and recovery). In order to ensure maximum reproducibility in gel pore size and to obtain a fully polymerized and non-restrictive large pore gel for a PAGE run, the polyacrylamide gel is polymerized for a time period of 69 hr at room temperature (RT). The exothermic heat generated by the polymerization processes is dissipated constantly while the temperature may rise rapidly to over 75 °C in the first minutes, after which it falls slowly.[17] After 69 hr the gel has reached room temperature, and thus, is in its lowest energy state because the chemical reactions and the gelation are terminated. Gelation means that the solvent (water) gets immobilized within the polymer network by means of hydrogen bonds and also van der Waals forces. As a result, the prepared gel is homogeneous (in terms of homogeneous distribution of cross-links throughout the gel sample[18]), inherently stable and free of monomers or radicals. Fresh polyacrylamide gels are further hydrophilic, electrically neutral and do not bind proteins.[19] Sieving effects due to gravity-induced compression of the gel can be excluded for the same reasons. In a medium without molecular sieving properties a high-resolution can be expected.[20]
Before an electrophoretic run is started the prepared 4% T (total polymer content (T)), 2.67% C (cross-linker concentration (C)) gel is pre-run to equilibrate it. It is essentially non-sieving and optimal for electrophoresis of proteins greater than or equal to 200 ku (cf. agarose gel electrophoresis). Proteins migrate in it more or less on the basis of their free mobility.[21] For these reasons interactions of the gel with the biomolecules are negligibly low, and thus, the proteins separate cleanly and predictably at a polymerization time of 69 hr (see figure Electropherogram). The separated metalloproteins including biomolecules ranging from approximately < 1 ku to greater than 30 ku (e.g., metal chaperones, prions, metal transport proteins, amyloids, metalloenzymes, metallopeptides, metallothionein, phytochelatins) are not dissociated into apoproteins and metal cofactors.[22]
Reproducibility and recovery
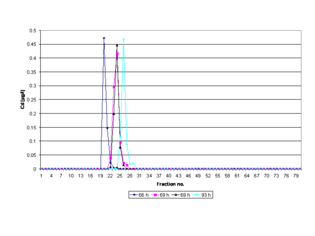
The bioactive structures (native or 3D conformation or shape) of the isolated protein molecules do not undergo any significant conformational changes. Thus, active metal cofactor-containing proteins can be isolated reproducibly in the same fractions after a PAGE run (see figure Electropherogram). A shifting peak in the respective electropherogram may indicate that the standardized time of gel polymerization (69 hr, RT) is not implemented in a PAGE experiment. A lower deviation of the standardized polymerization time (< 69 hr) stands for incomplete polymerization, and thus, for inherent instability due to gel softening during the cross-linking of polymers as the material reaches swelling equilibrium,[23] whereas exceeding this time limit (> 69 hr) is an indicator of gel aging.[24] The phenomenon of gel aging is closely connected to long-term viscosity decrease of aqueous polyacrylamide solutions[25] and increased swelling of hydrogels.[26]
Under standard conditions (69 hr), metalloproteins with different molecular mass ranges and isoelectric points have been recovered in biologically active form at a quantitative yield of more than 95%.[27] By preparative SDS polyacrylamide gel electrophoresis standard proteins (cytochrome c, aldolase, ovalbumin and bovine serum albumin) with molecular masses of 14-66 ku can be recovered with an average yield of about 73,6%.[28] Preparative isotachophoresis (ITP) is applied for isolating palladium-containing proteins with molecular masses of 362 ku (recovery: 67%) and 158 ku (recovery: 97%).[29]
Quantification and identification
Physiological concentrations (ppb-range) of Fe, Cu, Zn, Ni, Mo, Pd, Co, Mn, Pt, Cr, Cd and other metal cofactors can be identified and absolutely quantified in an aliquot of a fraction by inductively coupled plasma mass spectrometry (ICP-MS)[30] or total reflection X-ray fluorescence (TXRF),[31] for example. In case of ICP-MS the structural information of the associated metallobiomolecules[32] is irreversibly lost due to ionization of the sample with plasma.[33] Another established high sensitive detection method for the determination of (trace) elements is graphite furnace atomic absorption spectrometry (GF-AAS) (see figure Electropherogram).[34] Because of high purity and optimized concentration of the separated metalloproteins, for example, therapeutic recombinant plant-made pharmaceuticals such as copper chaperone for superoxide dismutase (CCS) from medicinal plants, in a few specific PAGE fractions, the related structures of these analytes can be elucidated quantitatively by using solution NMR spectroscopy under non-denaturing conditions.[35]
Applications
Improperly folded metal proteins, for example, CCS or Cu/Zn-superoxide dismutase (SOD1) present in brain, blood or other clinical samples, are indicative of neurodegenerative diseases like Alzheimer's disease (AD) or amyotrophic lateral sclerosis (ALS).[36] Active CCS or SOD molecules contribute to intracellular homeostatic control of essential metal ions (e.g., Cu1+/2+, Zn2+, Fe2+/3+, Mn2+, Ni3+) in organisms, and thus, these biomolecules can balance pro-oxidative and antioxidative processes in the cytoplasm. Otherwise, free (loosely bound) transition metal ions take part in Fenton-like reactions in which deleterious hydroxyl radical is formed, which unrestrained would be destructive to proteins.[37] The loss of (active) CCS increases the amyloid-β production in neurons which, in turn, is a major pathological hallmark of AD.[38] Therefore, copper chaperone for superoxide dismutase is proposed to be one of the most promising biomarkers of Cu toxicity in these diseases.[39] CCS should be analysed primarily in blood because a meta-analysis of serum data showed that AD patients have higher levels of serum Cu than healthy controls.[40]
See also
- Electrophoresis
- Gel electrophoresis
- Protein electrophoresis
- Protein purification
- Metallome
References
- Seelert H, Krause F (2008). "Preparative isolation of protein complexes and other bioparticles by elution from polyacrylamide gels". Electrophoresis. 29 (12): 2617–36. doi:10.1002/elps.200800061. PMID 18494038.
- Swart C, Jakubowski N (2016). "Update on the status of metrology for metalloproteins". Journal of Analytical Atomic Spectrometry. 31 (9): 1756–65. doi:10.1039/C6JA00181E.
- Finney LA, O'Halloran TV (2003). "Transition metal speciation in the cell: insights from the chemistry of metal ion receptors". Science. 300 (5621): 931–6. Bibcode:2003Sci...300..931F. doi:10.1126/science.1085049. PMID 12738850.
- Cerchiaro G, Manieri TM, Bertuchi FR (2013). "Analytical methods for copper, zinc and iron quantification in mammalian cells". Metallomics. 5 (10): 1336–45. doi:10.1039/c3mt00136a. PMID 23925479.
- Garfin DE (1990). Isoelectric focusing. Methods in Enzymology. 182. pp. 459–77. doi:10.1016/0076-6879(90)82037-3. ISBN 9780121820831. PMID 2314254.
- Youn HD, Kim EJ, Roe JH, Hah YC, Kang SO (1996). "A novel nickel-containing superoxide dismutase from Streptomyces spp". Biochemical Journal. 318 (Pt 3): 889–96. doi:10.1042/bj3180889. PMC 1217701. PMID 8836134.
- Suck R, Petersen A, Weber B, Fiebig H, Cromwell O (2004). "Analytical and preparative native polyacrylamide gel electrophoresis: investigation of the recombinant and natural major grass pollen allergen Phl p 2". Electrophoresis. 25 (1): 14–9. doi:10.1002/elps.200305697. PMID 14730563.
- Bae SH, Harris AG, Hains PG, Chen H, Garfin DE, Hazell SL, Paik YK, Walsh BJ, Cordwell SJ (2003). "Strategies for the enrichment and identification of basic proteins in proteome projects". Proteomics. 3 (5): 569–79. doi:10.1002/pmic.200300392. PMID 12748937.
- Kastenholz B (2006). "Comparison of the electrochemical behavior of the high molecular mass cadmium proteins in Arabidopsis thaliana and in vegetable plants on using preparative native continuous polyacrylamide gel electrophoresis (PNC-PAGE)". Electroanalysis. 18 (1): 103–6. doi:10.1002/elan.200403344.
- McLellan T (1982). "Electrophoresis buffers for polyacrylamide gels at various pH". Analytical Biochemistry. 126 (1): 94–9. doi:10.1016/0003-2697(82)90113-0. PMID 7181120.
- Kastenholz B, Garfin DE (2010). "Isolation of acidic, basic and neutral metalloproteins by QPNC-PAGE". Nature Precedings: 1–4. doi:10.1038/npre.2010.4617.1.
- Gordon AH (1969). Electrophoresis of proteins in polyacrylamide and starch gels (Part I, Chapter 2 Acrylamide gel). Laboratory Techniques in Biochemistry and Molecular Biology. 1. pp. 34–45. doi:10.1016/S0075-7535(08)70324-3. ISBN 9780444533418.
- Woolley P (1987). "Thermal instability of electrophoresis gels". Electrophoresis. 8 (8): 339–45. doi:10.1002/elps.1150080802.
- Gelfi C, Righetti PG (1981). "Polymerization kinetics of polyacrylamide gels II. Effect of temperature". Electrophoresis. 2 (4): 220–28. doi:10.1002/elps.1150020405.
- Chen B, Chrambach A (1979). "Estimation of polymerization efficiency in the formation of polyacrylamide gel, using continuous optical scanning during polymerization". Journal of Biochemical and Biophysical Methods. 1 (2): 105–16. doi:10.1016/0165-022X(79)90017-4. PMID 551105.
- Bonaventura C, Bonaventura J, Stevens R, Millington D (1994). "Acrylamide in polyacrylamide gels can modify proteins during electrophoresis". Analytical Biochemistry. 222 (1): 44–8. doi:10.1006/abio.1994.1451. PMID 7856869.
- Wheatley MA, Phillips CR (1983). "Temperature effects during polymerization of polyacrylamide gels used for bacterial cell immobilization". Biotechnology and Bioengineering. 25 (2): 623–6. doi:10.1002/bit.260250228. PMID 18548679.
- Kizilay MY, Okay O (2003). "Effect of hydrolysis on spatial inhomogeneity in poly(acrylamide) gels of various crosslink densities". Polymer. 44 (18): 5239–50. doi:10.1016/S0032-3861(03)00494-4.
- Garfin DE (2009). "25th Annual Meeting of the American Electrophoresis Society". Expert Review of Proteomics. 6 (3): 239–41. doi:10.1586/epr.09.18. PMID 19489696.
- Hjertén S (1963). ""Molecular-sieve" electrophoresis in cross-linked polyacrylamide gels". Journal of Chromatography A. 11: 66–70. doi:10.1016/S0021-9673(01)80870-0. PMID 13954823.
- Garfin DE (2009) [1990]. Chapter 29 one-dimensional gel electrophoresis. Methods in Enzymology. 463. pp. 497–513. doi:10.1016/S0076-6879(09)63029-9. ISBN 978-0-12-374536-1. PMID 19892189.
- Fitri N, Kastenholz B, Buchari B, Amran MB, Warganegara FM (2008). "Molybdenum speciation in raw phloem sap of castor bean". Analytical Letters. 41 (10): 1773–84. doi:10.1080/00032710802162442.
- Damljanović V, Lagerholm BC, Jacobson K (2005). "Bulk and micropatterned conjugation of extracellular matrix proteins to characterized polyacrylamide substrates for cell mechanotransduction assays". BioTechniques. 39 (6): 847–51. doi:10.2144/000112026. PMID 16382902.
- Stejskal J, Gordon M, Torkington JA (1980). "Collapse of polyacrylamide gels". Polymer Bulletin. 3 (11): 621–5. doi:10.1007/BF01135333.
- Kulicke WM, Kniewske R, Klein J (1982). "Preparation, characterization, solution properties and rheological behaviour of polyacrylamide". Progress in Polymer Science. 8 (4): 373–468. doi:10.1016/0079-6700(82)90004-1.
- Yoon J, Cai S, Suo Z, Hayward RC (2010). "Poroelastic swelling kinetics of thin hydrogel layers: comparison of theory and experiment". Soft Matter. 6 (23): 6004–12. Bibcode:2010SMat....6.6004Y. doi:10.1039/C0SM00434K.
- Kastenholz B (2006). "Important contributions of a new quantitative preparative native continuous polyacrylamide gel electrophoresis (QPNC-PAGE) procedure for elucidating metal cofactor metabolisms in protein-misfolding diseases – a theory". Protein and Peptide Letters. 13 (5): 503–8. doi:10.2174/092986606776819637. PMID 16800806.
- Ohhashi T, Moritani C, Andoh H, Satoh S, Ohmori S, Lottspeich F, Ikeda M (1991). "Preparative high-yield electroelution of proteins after separation by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and its application to analysis of amino acid sequences and to raise antibodies". Journal of Chromatography. 585 (1): 153–9. doi:10.1016/0021-9673(91)85069-r. PMID 1666109.
- Weber G, Messerschmidt J, von Bohlen A, Kastenholz B, Günther K (2004). "Improved separation of palladium species in biological matrices by using a combination of gel permeation chromatography and isotachophoresis". Electrophoresis. 25 (12): 1758–64. doi:10.1002/elps.200305833. PMID 15213973.
- Rašovský P (2011). "Utilisation of column gel eletrophoresis for on-line connection to ICP-MS for metalloproteomics". Archive of Thesis, Masaryk University, Brno.
- Pessanha S, Carvalho ML, Becker M, von Bohlen A (2010). "Quantitative determination on heavy metals in different stages of wine production by total reflection X-ray fluorescence and energy dispersive X-ray fluorescence: comparison on two vineyards". Spectrochimica Acta Part B Atomic Spectroscopy. 65 (6): 504–7. Bibcode:2010AcSpe..65..504P. doi:10.1016/j.sab.2010.04.003.
- Jakubowski N, Lobinski R, Moens L (2004). "Metallobiomolecules. The basis of life, the challenge of atomic spectroscopy". Journal of Analytical Atomic Spectrometry. 19 (1): 1–4. doi:10.1039/B313299B.
- Mounicou S, Szpunar J, Lobinski R (2009). "Metallomics: the concept and methodology". Chemical Society Reviews. 38 (4): 1119–38. doi:10.1039/B713633C. PMID 19421584.
- Lin TW, Huang SD (2001). "Direct and simultaneous determination of copper, chromium, aluminum, and manganese in urine with a multielement graphite furnace atomic absorption spectrometer". Analytical Chemistry. 73 (17): 4319–25. doi:10.1021/ac010319h. PMID 11569826.
- Kastenholz B, Garfin DE (2009). "Medicinal plants: a natural chaperones source for treating neurological disorders". Protein and Peptide Letters. 16 (2): 116–20. doi:10.2174/092986609787316234. PMID 19200033.
- Schümann K, Classen HG, Dieter HH, König J, Multhaup G, Rükgauer M, Summer KH, Bernhardt J, Biesalski HK (2002). "Hohenheim consensus workshop: copper". European Journal of Clinical Nutrition. 56 (6): 469–83. doi:10.1038/sj.ejcn.1601315. PMID 12032645.
- Robinson NJ, Winge DR (2010). "Copper metallochaperones". Annual Review of Biochemistry. 79: 537–62. doi:10.1146/annurev-biochem-030409-143539. PMC 3986808. PMID 20205585.
- Gray EH, De Vos KJ, Dingwall C, Perkinton MS, Miller CC (2010). "Deficiency of the copper chaperone for superoxide dismutase increases amyloid-β production". Journal of Alzheimer's Disease. 21 (4): 1101–5. doi:10.3233/JAD-2010-100717. PMC 3023902. PMID 20693630.
- Pal A (2014). "Copper toxicity induced hepatocerebral and neurodegenerative diseases: an urgent need for prognostic biomarkers". Neurotoxicology. 40: 97–101. doi:10.1016/j.neuro.2013.12.001. PMID 24342654.
- Bucossi S, Ventriglia M, Panetta V, Salustri C, Pasqualetti P, Mariani S, Siotto M, Rossini PM, Squitti R (2011). "Copper in Alzheimer's disease: a meta-analysis of serum, plasma, and cerebrospinal fluid studies". Journal of Alzheimer's Disease. 24 (1): 175–85. doi:10.3233/JAD-2010-101473. PMID 21187586.
External links
- Chapters on 1-D / 2-D Gel Electrophoresis, and Isoelectric Focusing are provided by AES Electrophoresis Society Learning Center: Foci
- Protein Purification Protocols by The Hebrew University of Jerusalem