Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) is a physical technique used to determine the thermodynamic parameters of interactions in solution. It is most often used to study the binding of small molecules (such as medicinal compounds) to larger macromolecules (proteins, DNA etc.).[1] It consists of two cells which are enclosed in an adiabatic jacket. The compounds to be studied are placed in the sample cell, while the other cell, the reference cell, is used as a control and contains the buffer in which the sample is dissolved.
| Acronym | ITC |
|---|---|
| Classification | Thermal analysis |
| Manufacturers | TA Instruments, Microcal/Malvern Instruments |
| Other techniques | |
| Related | Isothermal microcalorimetry Differential scanning calorimetry |
Thermodynamic measurements
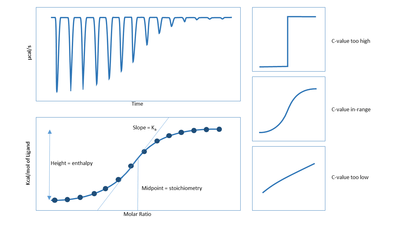
ITC is a quantitative technique that can determine the binding affinity (), enthalpy changes (), and binding stoichiometry () of the interaction between two or more molecules in solution. From these initial measurements, Gibbs free energy changes () and entropy changes () can be determined using the relationship:
(where is the gas constant and is the absolute temperature).
For accurate measurements of binding affinity, the curve of the thermogram must be sigmoidal. The profile of the curve is determined by the c-value, which is calculated using the equation:
where is the stoichiometry of the binding, is the association constant and is the concentration of the molecule in the cell.
The instrument
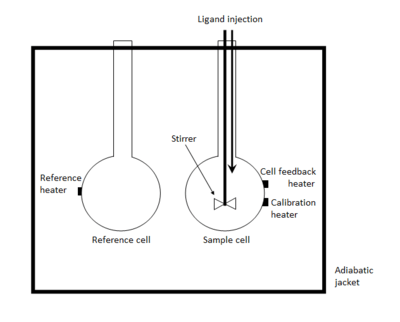
An isothermal titration calorimeter is composed of two identical cells made of a highly efficient thermally conducting and chemically inert material such as Hastelloy alloy or gold, surrounded by an adiabatic jacket.[2] Sensitive thermopile/thermocouple circuits are used to detect temperature differences between the reference cell (filled with buffer or water) and the sample cell containing the macromolecule. Prior to addition of ligand, a constant power (<1 mW) is applied to the reference cell. This directs a feedback circuit, activating a heater located on the sample cell.[3] During the experiment, ligand is titrated into the sample cell in precisely known aliquots, causing heat to be either taken up or evolved (depending on the nature of the reaction). Measurements consist of the time-dependent input of power required to maintain equal temperatures between the sample and reference cells.
In an exothermic reaction, the temperature in the sample cell increases upon addition of ligand. This causes the feedback power to the sample cell to be decreased (remember: a reference power is applied to the reference cell) in order to maintain an equal temperature between the two cells. In an endothermic reaction, the opposite occurs; the feedback circuit increases the power in order to maintain a constant temperature (isothermic/isothermal operation).
Observations are plotted as the power needed to maintain the reference and the sample cell at an identical temperature against time. As a result, the experimental raw data consists of a series of spikes of heat flow (power), with every spike corresponding to one ligand injection. These heat flow spikes/pulses are integrated with respect to time, giving the total heat exchanged per injection. The pattern of these heat effects as a function of the molar ratio [ligand]/[macromolecule] can then be analysed to give the thermodynamic parameters of the interaction under study. Degassing samples is often necessary in order to obtain good measurements as the presence of gas bubbles within the sample cell will lead to abnormal data plots in the recorded results. The entire experiment takes place under computer control.
Application in drug discovery
ITC is one of the latest techniques to be used in characterizing binding affinity of ligands for proteins. It is typically used as a secondary screening technique in high throughput screening. ITC is particularly useful as it gives not only the binding affinity, but also the thermodynamics of the binding. This thermodynamic characterization allows for further optimization of compounds.[4]
See also
- Differential scanning calorimetry
- Dual polarisation interferometry
- Pressure perturbation calorimetry
- Surface plasmon resonance
References
- Pierce, Michael M.; Raman, C.S.; Nall, Barry T. (1999). "Isothermal Titration Calorimetry of Protein–Protein Interactions". Methods. 19 (2): 213–221. CiteSeerX 10.1.1.385.1426. doi:10.1006/meth.1999.0852. PMID 10527727.
- O’Brien, R., Ladbury, J.E. and Chowdry B.Z. (2000) Isothermal titration calorimetry of biomolecules. Chapter 10 in Protein-Ligand interactions: hydrodynamics and calorimetry Ed. Harding, S.E. and Chowdry, B.Z, Oxford University Press. ISBN 0-19-963746-6
- VP-ITC Instruction Manual (2001). Microcal Inc., Northampton, MA. http://www.microcalorimetry.com
- Desphande, T.Isothermal Titration Calorimetry (ITC): Application in Drug Discovery