Ebolavirus
The genus Ebolavirus (/iˈboʊlə/- or /əˈboʊləˌvaɪrəs/; ee-BOH-lə- or ə-BOH-lə-VY-rəs)[1][2][3] is a virological taxon included in the family Filoviridae (filament-shaped viruses), order Mononegavirales.[3] The members of this genus are called ebolaviruses,[3] and encode their genome in the form of single-stranded negative-sense RNA.[4] The six known virus species are named for the region where each was originally identified: Bundibugyo ebolavirus, Reston ebolavirus, Sudan ebolavirus, Taï Forest ebolavirus (originally Côte d'Ivoire ebolavirus), Zaire ebolavirus, and Bombali ebolavirus. The last is the most recent species to be named and was isolated from Angolan free-tailed bats in Sierra Leone.[5]
| Ebolavirus | |
|---|---|
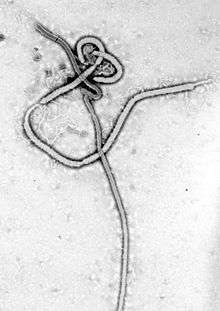 | |
| Ebola virus under transmission electron microscope | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Filoviridae |
| Genus: | Ebolavirus |
| Type species | |
| Zaire ebolavirus | |
| Species | |
| |
Each species of the genus Ebolavirus has one member virus, and four of these cause Ebola virus disease (EVD) in humans, a type of hemorrhagic fever having a very high case fatality rate. The Reston virus has caused EVD in other primates.[6][7] Zaire ebolavirus is the type species (reference or example species) for Ebolavirus, has the highest mortality rate of the ebolaviruses, and is responsible for the largest number of outbreaks of the six known species of the genus, including the 1976 Zaire outbreak and the outbreak with the most deaths (2014).[8]
Ebolaviruses were first described after outbreaks of EVD in southern Sudan in June 1976 and in Zaire in August 1976.[9][10] The name Ebolavirus is derived from the Ebola River in Zaire (now the Democratic Republic of the Congo), near the location of the 1976 outbreak,[10] and the taxonomic suffix -virus (denoting a viral genus).[3] This genus was introduced in 1998 as the "Ebola-like viruses".[11][12] In 2002, the name was changed to Ebolavirus[13][14] and in 2010, the genus was emended.[3] Ebolaviruses are closely related to marburgviruses.
Hosts of the Ebolavirus
Researchers have found evidence of Ebola infection in three species of fruit bat. The bats show no symptoms of the disease, indicating that they may be the main natural reservoirs of the Ebolavirus. It is possible that there are other reservoirs and vectors. Understanding where the virus incubates between outbreaks and how it is transmitted between species will help protect humans and other primates from the virus.
The researchers found that bats of three species — Franquet's epauletted fruit bat (Epomops franqueti), the hammer-headed bat (Hypsignathus monstrosus) and the little collared fruit bat (Myonycteris torquata) — had either genetic material from the Ebola virus, known as RNA sequences, or evidence of an immune response to the disease. The bats showed no symptoms themselves.[15]
The Bombali ebolavirus (BOMV) was isolated from the little free-tailed bat (Chaerephon pumilus) and the Angolan free-tailed bat (Mops condylurus) in Sierra Leone.[5]
Entry pathway
The entry pathway that the virus uses is a key step in its cycle. Several pathways have been suggested for Ebolavirus such as phagocytosis and clathrin and caveolin mediated endocytosis. However, Nanbo et al. (2010) proved that neither of these pathways is actually used.[16]
They discovered that Ebolavirus uses macropinocytosis to enter the host cells. Induction of macropinocytosis leads to the formation of macropinocytosis-specific endosomes (macropinosomes), which are large enough to accommodate Ebola virions. This discovery was proven by the fact that Ebolavirus co-localizes with sorting nexin 5 (SNX5), which consists of a large family of peripheral membrane proteins that associate with newly formed macropinosomes.[16]
Then, internalized EBOV particles are transported to late endosomes and, there, co-localization with GTPase Rab7 (marker of late endosomes) is observed.[16]
Also, blocking the macropinocytosis pathway has been proven to stop Ebolavirus from entering the cells. Four different macropinocytosis specific inhibitors were tested: cytochalasin D (depolymerizing agent), wortmannin (Wort), LY-294002 (both are inhibitors of PI3K) and EIPA (5-(N-ethyl-N-isopropyl) amiloride), an inhibitor of the Na+/H+ exchanger specific for pinocytosis.[16]
Taxonomy notes
According to the rules for taxon naming established by the International Committee on Taxonomy of Viruses (ICTV), the name of the genus Ebolavirus is always to be capitalized, italicized, never abbreviated, and to be preceded by the word "genus". The names of its members (ebolaviruses) are to be written in lower case, are not italicized, and used without articles.[3]
Genus inclusion criteria
A virus of the family Filoviridae is a member of the genus Ebolavirus if[3]
- its genome has several gene overlaps
- its fourth gene (GP) encodes four proteins (sGP, ssGP, Δ-peptide, and GP1,2) using cotranscriptional editing to express ssGP and GP1,2 and proteolytic cleavage to express sGP and Δ-peptide
- peak infectivity of its virions is associated with particles ≈805 nm in length
- its genome differs from that of Marburg virus by ≥50% and from that of Zaire ebolavirus by <50% at the nucleotide level
- its virions show almost no antigenic cross reactivity with marburg virions
Classification
.jpg)
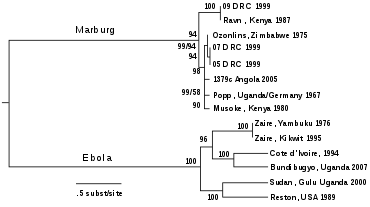
The genera Ebolavirus and Marburgvirus were originally classified as the species of the now-obsolete genus Filovirus. In March 1998, the Vertebrate Virus Subcommittee proposed in the International Committee on Taxonomy of Viruses (ICTV) to change the genus Filovirus to the family Filoviridae with two specific genera: Ebola-like viruses and Marburg-like viruses. This proposal was implemented in Washington, D.C., as of April 2001 and in Paris as of July 2002. In 2000, another proposal was made in Washington, D.C., to change the "-like viruses" to "-virus" resulting in today's Ebolavirus and Marburgvirus.[17][18]
The five characterised species of the genus Ebolavirus are:
- Zaire ebolavirus (ZEBOV)
- Also known simply as the Zaire virus, ZEBOV has the highest case-fatality rate, up to 90% in some epidemics, with an average case fatality rate of approximately 83% over 27 years. There have been more outbreaks of Zaire ebolavirus than of any other species. The first outbreak took place on 26 August 1976 in Yambuku.[19] Mabalo Lokela, a 44‑year-old schoolteacher, became the first recorded case. The symptoms resembled malaria, and subsequent patients received quinine. Transmission has been attributed to reuse of unsterilized needles and close personal contact. The virus is responsible for the 2014 West Africa Ebola virus outbreak, with the largest number of deaths to date.
- Sudan ebolavirus (SUDV)
- Like ZEBOV, SUDV emerged in 1976; it was at first assumed to be identical with ZEBOV.[20] SUDV is believed to have broken out first amongst cotton factory workers in Nzara, Sudan (now in South Sudan), in June 1976, with the first case reported as a worker exposed to a potential natural reservoir. Scientists tested local animals and insects in response to this; however, none tested positive for the virus. The carrier is still unknown. The lack of barrier nursing (or "bedside isolation") facilitated the spread of the disease. The average fatality rates for SUDV were 54% in 1976, 68% in 1979, and 53% in 2000 and 2001.
- Reston ebolavirus (RESTV)
- This virus was discovered during an outbreak of simian hemorrhagic fever virus (SHFV) in crab-eating macaques from Hazleton Laboratories (now Covance) in 1989. Since the initial outbreak in Reston, Virginia, it has since been found in nonhuman primates in Pennsylvania, Texas, and Siena, Italy. In each case, the affected animals had been imported from a facility in the Philippines,[21] where the virus has also infected pigs.[22] Despite its status as a Level‑4 organism and its apparent pathogenicity in monkeys, RESTV did not cause disease in exposed human laboratory workers.[23]
- Taï Forest ebolavirus (TAFV)
- Formerly known as "Côte d'Ivoire ebolavirus", it was first discovered among chimpanzees from the Tai Forest in Côte d'Ivoire, Africa, in 1994. Necropsies showed blood within the heart to be brown; no obvious marks were seen on the organs; and one necropsy displayed lungs filled with blood. Studies of tissues taken from the chimpanzees showed results similar to human cases during the 1976 Ebola outbreaks in Zaire and Sudan. As more dead chimpanzees were discovered, many tested positive for Ebola using molecular techniques. The source of the virus was believed to be the meat of infected western red colobus monkeys (Procolobus badius) upon which the chimpanzees preyed. One of the scientists performing the necropsies on the infected chimpanzees contracted Ebola. She developed symptoms similar to those of dengue fever approximately a week after the necropsy, and was transported to Switzerland for treatment. She was discharged from hospital after two weeks and had fully recovered six weeks after the infection.[24]
- Bundibugyo ebolavirus (BDBV)
- On November 24, 2007, the Uganda Ministry of Health confirmed an outbreak of Ebola in the Bundibugyo District. After confirmation of samples tested by the United States National Reference Laboratories and the CDC, the World Health Organization confirmed the presence of the new species. On 20 February 2008, the Uganda Ministry officially announced the end of the epidemic in Bundibugyo, with the last infected person discharged on 8 January 2008.[25] An epidemiological study conducted by WHO and Uganda Ministry of Health scientists determined there were 116 confirmed and probable cases of the new Ebola species, and that the outbreak had a mortality rate of 34% (39 deaths).[26]
Evolution
Rates of genetic change are 8*10−4 per site per year and are thus one fourth[27] as fast as influenza A in humans. Extrapolating backwards, Ebolavirus and Marburgvirus probably diverged several thousand years ago.[28] A study done in 1995 and 1996 found that the genes of Ebolavirus and Marburgvirus differed by about 55% at the nucleotide level, and at least 67% at the amino acid level. The same study found that the strains of Ebolavirus differed by about 37-41% across the nucleotide level and 34-43% across the amino acid level. The EBOV strain was found to have an almost 2% change in the nucleotide level from the original 1976 strain from the Yambuki outbreak and the strain from the 1995 Kikwit outbreak.[29] However, paleoviruses of filoviruses found in mammals indicate that the family itself is at least tens of millions of years old.[30]
Genus organization and common names
The genus Ebolavirus has been organized into five species; however, the nomenclature has proven somewhat controversial, with many authors continuing to use common names rather than species names when referring to these viruses.[3] In particular, the generic term "Ebola virus" is widely used to refer specifically to members of the species Zaire ebolavirus. Consequently, in 2010, a group of researchers recommended that the name "Ebola virus" be adopted for a subclassification[note 1] within the species Zaire ebolavirus and that similar common names be formally adopted for other Ebolavirus species.[3] In 2011, the International Committee on Taxonomy of Viruses (ICTV) rejected a proposal (2010.010bV) to formally recognize these names, as they do not designate names for subtypes, variants, strains, or other subspecies level groupings.[31] As such, the widely used common names are not formally recognized as part of the taxonomic nomenclature. In particular, "Ebola virus" does not have an official meaning recognized by ICTV, and rather they continue to use and recommend only the species designation Zaire ebolavirus.[32]
The threshold for putting isolates into different species is usually a difference of more than 30% at the nucleotide level, compared to the type strain. If a virus is in a given species but differs from the type strain by more than 10% at the nucleotide level, it is proposed that it be named as a new virus. As of 2019, none of the Ebolavirus species contain members divergent enough to receive more than one "virus" designation.[3]
| Species name (Abbreviation) | Virus common name (Abbreviation)[3] |
|---|---|
| Bombali ebolavirus | Bombali virus (BOMV) |
| Bundibugyo ebolavirus (BEBOV) | Bundibugyo virus (BDBV) |
| Reston ebolavirus (REBOV) | Reston virus (RESTV) |
| Sudan ebolavirus (SEBOV) | Sudan virus (SUDV) |
| Taï Forest ebolavirus (TEBOV; previously CIEBOV) | Taï Forest virus (TAFV) |
| Zaire ebolavirus (ZEBOV) | Ebola virus (EBOV) |
Research
A 2013 study isolated antibodies from fruit bats in Bangladesh, against Ebola Zaire and Reston viruses, thus identifying potential virus hosts and signs of the filoviruses in Asia.[33]
A recent alignment-free analysis of Ebola virus genomes from the current outbreak reveals the presence of three short DNA sequences that appear nowhere in the human genome, suggesting that the identification of specific species sequences may prove to be useful for the development of both diagnosis and therapeutics.[34]
Notes
- The Kuhn et al. 2010 proposal specifically suggested that "Ebola virus" be given a taxonomic rank of "Virus" within the species Zaire ebolavirus. In their proposal, an "Ebola virus" would be any member of species Zaire ebolavirus whose genome diverged from the type variant Zaire ebolavirus (Mayinga) by less than 10%. In general, the members of species Zaire ebolavirus are allowed to genetically diverge from the Mayinga type variant by up to 30%.[3] As a result, this proposal would make "Ebola virus" a subset of the species Zaire ebolavirus rather than a common name synonym. The distinction of treating "Ebola virus" as a subset of the species rather than as a synonym for the species is rarely used.
References
- American Heritage Dictionary
- Cambridge Advanced Learner's Dictionary
- Kuhn, J. H.; Becker, S.; Ebihara, H.; Geisbert, T. W.; Johnson, K. M.; Kawaoka, Y.; Lipkin, W. I.; Negredo, A. I.; Netesov, S. V.; Nichol, S. T.; Palacios, G.; Peters, C. J.; Tenorio, A.; Volchkov, V. E.; Jahrling, P. B. (2010). "Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations". Archives of Virology. 155 (12): 2083–103. doi:10.1007/s00705-010-0814-x. PMC 3074192. PMID 21046175.
- Kuhn, JH; Amarasinghe, GK; Basler, CF; Bavari, S; Bukreyev, A; Chandran, K; Crozier, I; Dolnik, O; Dye, JM; Formenty, PBH; Griffiths, A; Hewson, R; Kobinger, GP; Leroy, EM; Mühlberger, E; Netesov, SV; Palacios, G; Pályi, B; Pawęska, JT; Smither, SJ; Takada, A; Towner, JS; Wahl, V; ICTV Report, Consortium (June 2019). "ICTV Virus Taxonomy Profile: Filoviridae". The Journal of General Virology. 100 (6): 911–912. doi:10.1099/jgv.0.001252. PMID 31021739.
- "New Ebola species is reported for first time in a decade - STAT". STAT. 27 July 2018. Retrieved 28 July 2018.
- Spickler, Anna. "Ebolavirus and Marburgvirus Infections" (PDF).
- "About Ebola Virus Disease". Centers for Disease Control. Retrieved 18 October 2014.
- Kamorudeen, Ramat Toyin; Adedokun, Kamoru Ademola; Olarinmoye, Ayodeji Oluwadare (May 2020). "Ebola outbreak in West Africa, 2014 – 2016: Epidemic timeline, differential diagnoses, determining factors, and lessons for future response". Journal of Infection and Public Health: S1876034120304275. doi:10.1016/j.jiph.2020.03.014.
- "Home" (PDF). Archived from the original (PDF) on 2014-10-13. Retrieved 2014-10-07.
- Kalra S., Kelkar D., Galwankar S. C., Papadimos T. J., Stawicki S. P., Arquilla B., Hoey B. A., Sharpe R. P., Sabol D., Jahre J. A. The emergence of Ebola as a global health security threat: From 'lessons learned' to coordinated multilateral containment efforts. J Global Infect Dis [serial online] 2014 [cited 2015 Mar 1]; 6:164–77.
- Netesov, S. V.; Feldmann, H.; Jahrling, P. B.; Klenk, H. D.; Sanchez, A. (2000). "Family Filoviridae". In van Regenmortel, M. H. V.; Fauquet, C. M.; Bishop, D. H. L.; Carstens, E. B.; Estes, M. K.; Lemon, S. M.; Maniloff, J.; Mayo, M. A.; McGeoch, D. J.; Pringle, C. R.; Wickner, R. B. (eds.). Virus Taxonomy—Seventh Report of the International Committee on Taxonomy of Viruses. San Diego, U.S.: Academic Press. pp. 539–48. ISBN 978-0-12-370200-5{{inconsistent citations}}CS1 maint: ref=harv (link)
- Pringle, C. R. (1998). "Virus taxonomy-San Diego 1998". Archives of Virology. 143 (7): 1449–59. doi:10.1007/s007050050389. PMID 9742051. S2CID 13229117.
- Feldmann, H.; Geisbert, T. W.; Jahrling, P. B.; Klenk, H.-D.; Netesov, S. V.; Peters, C. J.; Sanchez, A.; Swanepoel, R.; Volchkov, V. E. (2005). "Family Filoviridae". In Fauquet, C. M.; Mayo, M. A.; Maniloff, J.; Desselberger, U.; Ball, L. A. (eds.). Virus Taxonomy – Eighth Report of the International Committee on Taxonomy of Viruses. San Diego, U.S.: Elsevier/Academic Press. pp. 645–653. ISBN 978-0-12-370200-5{{inconsistent citations}}CS1 maint: ref=harv (link)
- Mayo, M. A. (2002). "ICTV at the Paris ICV: results of the plenary session and the binomial ballot". Archives of Virology. 147 (11): 2254–60. doi:10.1007/s007050200052. S2CID 43887711.
- "Fruit Bats Likely Hosts of Deadly Ebola Virus". news.nationalgeographic.com. Retrieved 2017-05-14.
- Nanbo, Asuka; Masaki, Imai; Shinji, Watanabe; Noda, Takeshi; Takahash, Kei; Neuman, Gabriele; Halfmann, Peter; Kawaoka, Yoshihiro (23 September 2010). "Ebolavirus Is Internalized into Host Cells via Macropinocytosis in a Viral Glycoprotein-Dependent Manner". PLOS Pathogens. 6 (9): e1001121. doi:10.1371/journal.ppat.1001121. PMC 2944813. PMID 20886108.
- Büchen-Osmond, Cornelia (2006-04-25). "ICTVdB Virus Description – 01.025.0.02. Ebolavirus". International Committee on Taxonomy of Viruses. Archived from the original on February 14, 2009. Retrieved 2009-06-02.
- "Virus Taxonomy: 2013 Release". International Committee on Taxonomy of Viruses. 2013. Retrieved 2014-10-31.
- Isaacson, M; Sureau, P; Courteille, G; Pattyn, SR. "Clinical Aspects of Ebola Virus Disease at the Ngaliema Hospital, Kinshasa, Zaire, 1976". Ebola Virus Haemorrhagic Fever: Proceedings of an International Colloquium on Ebola Virus Infection and Other Haemorrhagic Fevers Held in Antwerp, Belgium, 6–8 December 1977. Retrieved 2016-01-02.
- Feldmann, H.; Geisbert, T. W. (2011). "Ebola haemorrhagic fever". The Lancet. 377 (9768): 849–862. doi:10.1016/S0140-6736(10)60667-8. PMC 3406178. PMID 21084112.
- Special Pathogens Branch CDC (2008-01-14). "Known Cases and Outbreaks of Ebola Hemorrhagic Fever". Center for Disease Control and Prevention. Archived from the original on 2008-08-29. Retrieved 2008-08-02.
- McNeil Jr, Donald G. (2009-01-24). "Pig-to-Human Ebola Case Suspected in Philippines". New York Times. Retrieved 2009-01-26.
- McCormick & Fisher-Hoch 1999, p. 300
- Waterman, Tara (1999). Ebola Cote D'Ivoire Outbreaks. Stanford University. Archived from the original on 2008-02-16. Retrieved 2009-05-30.
- "End of Ebola outbreak in Uganda" (Press release). World Health Organization. 2008-02-20.
- Wamala, J; Lukwago, L; Malimbo, M; Nguku, P; Yoti, Z; Musenero, M; Amone, J; Mbabazi, W; Nanyunja, M; Zaramba, S; Opio, A; Lutwama, J; Talisuna, A; Okware, I (2010). "Ebola Hemorrhagic Fever Associated with Novel Virus Strain, Uganda, 2007–2008". Emerging Infectious Diseases. 16 (7): 1087–92. doi:10.3201/eid1607.091525. PMC 3321896. PMID 20587179.CS1 maint: ref=harv (link)
- Gire, S. K.; Goba, A.; Andersen, K. G.; Sealfon, R. S. G.; Park, D. J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; Wohl, S.; Moses, L. M.; Yozwiak, N. L.; Winnicki, S.; Matranga, C. B.; Malboeuf, C. M.; Qu, J.; Gladden, A. D.; Schaffner, S. F.; Yang, X.; Jiang, P.-P.; Nekoui, M.; Colubri, A.; Coomber, M. R.; Fonnie, M.; Moigboi, A.; Gbakie, M.; Kamara, F. K.; Tucker, V.; et al. (2014). "Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak". Science. 345 (6202): 1369–1372. Bibcode:2014Sci...345.1369G. doi:10.1126/science.1259657. PMC 4431643. PMID 25214632.
- Suzuki, Y; Gojobori, T. (1997). "The origin and evolution of Ebola and Marburg viruses". Molecular Biology and Evolution. 14 (8): 800–6. doi:10.1093/oxfordjournals.molbev.a025820. PMID 9254917.
- Sanchez, Anthony; Trappier, Sam; Mahy, Brian; Peters, Clarence; Nichol, Stuart (April 1996). "The virion glycoproteins of Ebola are encoded in two reading frames and are expressed through transcriptional editing". Proceedings of the National Academy of Sciences of the United States of America. 93 (8): 3602–3607. Bibcode:1996PNAS...93.3602S. doi:10.1073/pnas.93.8.3602. PMC 39657. PMID 8622982.
- Taylor, D.; Leach, R.; Bruenn, J. (2010). "Filoviruses are ancient and integrated into mammalian genomes". BMC Evolutionary Biology. 10: 193. doi:10.1186/1471-2148-10-193. PMC 2906475. PMID 20569424.
- "Replace the species name Lake Victoria marburgvirus with Marburg marburgvirus in the genus Marburgvirus".
- International Committee on Taxonomy of Viruses. "Virus Taxonomy: 2013 Release".
- Kevin J. Olival, Ariful Islam, Meng Yu, Simon J. Anthony, Jonathan H. Epstein, Shahneaz Ali Khan, Salah Uddin Khan, Gary Crameri, Lin-Fa Wang, W. Ian Lipkin, Stephen P. Luby, and Peter Daszak (2013). "Ebola Virus Antibodies in Fruit Bats, Bangladesh". Emerging Infectious Diseases. 19 (2): 270–3. doi:10.3201/eid1902.120524. PMC 3559038. PMID 23343532.CS1 maint: uses authors parameter (link)
- Raquel M. Silva, Diogo Pratas, Luísa Castro, Armando J. Pinho, Paulo J. S. G. Ferreira (2015). "Three minimal sequences found in Ebola virus genomes and absent from human DNA". Bioinformatics. 31 (15): 2421–2425. doi:10.1093/bioinformatics/btv189. PMC 4514932. PMID 25840045.CS1 maint: uses authors parameter (link)
External links
| Wikimedia Commons has media related to Ebolavirus. |
| Wikispecies has information related to Ebolavirus |