NEDD9
Neural precursor cell expressed developmentally down-regulated protein 9 (NEDD-9) is a protein that in humans is encoded by the NEDD9 gene.[5] NEDD-9 is also known as enhancer of filamentation 1 (EF1), CRK-associated substrate-related protein (CAS-L), and Cas scaffolding protein family member 2 (CASS2). An important paralog of this gene is BCAR1.
Discovery
In 1992, Kumar, et al., first described a sequence tag corresponding to the NEDD9 3′ untranslated region based on the cloning of a group of genes predominantly expressed in the brain of embryonic, but not adult mice, a group of genes designated neural precursor cell expressed, developmentally down-regulated.[6] In 1996, two groups independently described the complete sequence of the NEDD9 gene, and provided initial functional analysis of NEDD9 protein. Law et al. overexpressed a human cDNA library in S. cerevisiae, and screened for genes that simultaneously affected cell cycle and cell polarity controls, inducing a filamentous yeast budding phenotype, and thus identified the HEF1 protein (Human Enhancer of Filamentation 1).[7] This study identified HEF1/NEDD9 as an interactive partner for focal adhesion kinase (FAK), connecting it to integrin signaling. Separately, Minegishi et al. cloned the gene encoding a protein hyperphosphorylated following ligation of β1-integrins in T cells and hypothesized to play a role in the process of T cell costimulation, designating this gene Cas-L (Crk-associated substrate-related protein, Lymphocyte type).[8]
Gene
The genomic coordinates of the NEDD9 gene are 6:11,183,530-11,382,580 in the GRCh37 assembly, or 6:11,183,298-11,382,348 in the GRCh38 assembly. The gene is on the minus strand. The cytogenetic location is 6p25-p24, based on the nomenclature developed by the Human Genome Organization (HUGO) gene nomenclature committee (HGNC). NEDD9 is the HGNC approved symbol. Official IDs are 7733 (HGNC), 4739 (Entrez Gene), and ENSG00000111859 (Ensembl). CAS-L, CASL, HEF1, dJ49G10.2, dJ761I2.1, CAS2, CASS2 are alias symbols. The NEDD9 gene is conserved in Rhesus monkeys, dogs, cows, mice, rats, chickens, zebrafish, and frogs. In vertebrates, it is a member of a 4-gene family, with the other paralogous genes known as BCAR1 (p130Cas), EFS (Sin), and CASS4 (HEPL)
The NEDD9 promoter has 2 transcriptional start sites. The transcript variants NM_006403.3 and NM_001142393.1 encode proteins that have distinct N-termini (MKYK and MWTR, respectively). In mouse, the two alternative first exons are MKYK and MWAR. Their function is not known. NM_001142393 initiates translation at an upstream location compared to NM_006403.3, but both transcripts have 7 exons. Shorter transcripts with missing exons or an alternative 3' terminal exon have been detected in various studies; however, their role in the cell is unclear.
The 5' region of the NEDD9 promoter is regulated by all-trans retinoic acid (ATRA), and contains a retinoic acid response element (RARE) that is specifically bound by a retinoid X receptor (RXR)/retinoic acid receptor (RAR) heterodimer.[9][10][11] NEDD9 is also induced by the environmental pollutant dioxin, based on regulation through the aryl hydrocarbon receptor (AhR).[12] One study has found NEDD9 repressed by estrogen, based on binding of the SAFB1 co-repressor.[13] NEDD9 is induced by Wnt signaling in colon cancer, based on binding to T-cell factor (TCF) factors in the promoter region.[14] NEDD9 is induced by hypoxia and loss of VHL, based on binding of hypoxia-induced factor (HIF) transcription factors to the NEDD9 promoter.[15][16][17] Prostaglandin E2 induces NEDD9 transcription.[18] The Fox transcription factor Forkhead box C1 (FoxC1)[19] and PAX5 transcription factor [20] have been reported to induce NEDD9 transcription. TGF-beta induces NEDD9 transcription.[21] Based on inspection of sequence, the NEDD9 promoter also has potential binding sites for a number of additional transcription factors, including STAT5A and NF-kappa B.
In the 3'UTR of NEDD9 is a match to positions 2-8 of mature miR-145. NEDD9-binding regions in the miR-145 locus would allow the direct binding of the NEDD9 3'UTR to the genomic region of miR-145, and some studies suggests this miR regulates NEDD9 in glioblastoma [22] prostate cancer,[23] and renal cell carcinoma cells.[24] A non-coding RNA, named B2, extending from 10 kb upstream of NEDD9 exon 1 to exon 4 has been described, but the functional role for this ncRNA is not yet clear.[25] NEDD9 is highly expressed in the embryonal brain,[26] and in numerous tissues in the embryo and adult organism. Elevated expression is associated with cancer, as discussed below.
Protein family
NEDD9 is a member of the CAS (Crk-associated substrate) protein family, which has 4 members in vertebrates. The other paralogous genes are known BCAR1 (p130Cas),[27] EFS (Sin),[28][29] and CASS4 (HEPL).[30] There is no detectable NEDD9-related gene in bacteria, yeast, or C. elegans. A single family member exists in D. Melanogaster, termed DCas.[31][32]
Structure
In humans, NEDD9 is 834 amino acids long. NEDD9 is a noncatalytic scaffolding protein that contains docking sites for proteins involved in multiple signal transduction pathways, regulating magnitude and duration of cell signaling cascades [33][34][35][36] The overall structure of NEDD9 is represented graphically in Figure 1.
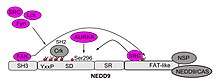
These domains include:
- SH3 domain
- This highly conserved N-terminal domain mediates NEDD9 binding to the polyproline motifs of a number of important interacting proteins, with some well-studied partners being FAK[7] and the related PYK2/RAFTK kinase,[37] C3G,[38] PTP-PEST,[39] PTP1B [40] and CIZ.[41]
- Substrate domain (SD)
- This unstructured region contains multiple YxxP motifs, which are phosphorylated by src family kinases (such as FYN, LCK and SRC) to create binding sites for proteins with SH2 domains, such as Crk.[8] Phosphorylation of these motifs can be activated by mechanical forces such as cytoskeletal stretch.[42] Other phosphorylation events in this region are imposed by the kinase Aurora-A, which phosphorylates residue S296, for processes related to cell cycle control.[43]
- Serine rich (SR) region
- The SR region likely folds into a 4-helix bundle, based on substantial predicted homology to BCAR1, for which the structure has been solved.[44]
- Focal adhesion targeting (FAT) domain
- The FAT-like C-terminal domain[45] is highly conserved in focal adhesion proteins, and sufficient for localizing focal adhesion kinase (FAK) to focal adhesions.[46] It forms a four-helix bundle structure and implicated in interaction with NSP proteins (novel SH2-containing protein family),[47][48] and other proteins such as the Id family of helix-loop-helix proteins.[49]
In terms of post-translational modifications, NEDD9 is subject to significant phosphorylation based on growth conditions. In most actively growing adherent cells, NEDD9 migrates as a doublet of 115 and 105 kDa. Serine/threonine hyper-phosphorylated p115 NEDD9 is more common in G2/M phase cells,[50] suggesting these modifications are associated with increased localization to centrosome and mitotic spindle. One study indicated the conversion of p115 into p105 is activated by cell detachment through cytoskeletal regulation of phosphatase PP2A,[51] although other work has found conflicting results.[52]
Synthesis and degradation
NEDD9 is present throughout cell cycle, but most abundant in G2/M phase cells.[50] NEDD9 is subject to both caspase cleavage and proteasomal degradation.[34][35] In conditions of cell detachment, and particularly in early stages of anoikis or apoptosis, NEDD9 is rapidly cleaved by caspases 3 and/or 7 at a DLVD site (residue 363), and at a DDYD site (residue 630) [53] to form N-terminal 55 KDa and C-terminal 28 KDa fragments forms. This cleavage is prevented by focal adhesion formation, which suggests NEDD9 as a sensor of altered adhesion states.[50][54] Overexpression of p28 in cells causes cellular rounding and detachment, and induces apoptosis,[54] probably because of a dominant-negative effect on survival-promoting signaling complexes at focal adhesions. Together this data suggests that production of different NEDD9 posttranslational modifications is regulated by cell de/attachment, which, in turn, allows regulation of NEDD9 turnover and participation in distinct cellular processes.
P115 is the primary target for proteasomal degradation of NEDD9.[51] Proteasomal degradation of NEDD9 is triggered by a number of stimuli, including induction of TGF-beta signaling.[55] An effector of the TGFbeta receptor, Smad3, may interact directly with APC subunit APC10 and thus recruit the APC complex. CDH1 subunit of the APC complex recognizes NEDD9 and regulates ubiquitination and subsequent degradation of NEDD9.[56] NEDD9 is also degraded by the proteasome at the end of mitosis, following completion of activities with Aurora-A that support mitotic progression.[50]
Tissue distribution and intracellular localization
In interphase cells, the majority of NEDD9 localizes to focal adhesions. However, some of the protein is also cytoplasmic, and small pools localize to the centrosome [43] and the basal body of cilia.[57] At mitotic entry NEDD9 moves along mitotic spindle, eventually localizing at midbody at cytokinesis.[43]
Function
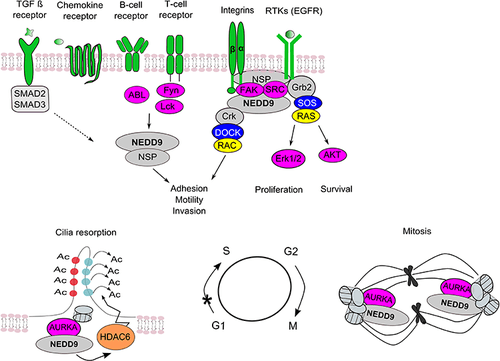
NEDD9 is an intermediate in a number of important signaling pathways relevant to the cellular processes of proliferation, survival, migration, and others (see figure to the right).[33][34][35]
Integrin, FAK/RAFTK, and SRC kinases
Integrin signaling, which control cell movement, spreading and adhesion to extracellular matrix (ECM), and survival, is the best established signaling pathway for NEDD9. Integrins are transmembrane proteins that nucleate focal adhesions, structures that provide bi-directional signaling between ECM and actin cytoskeleton. NEDD9 stabilizes formation and regulates turnover of focal adhesions, influencing cell motility and the invasion and metastasis of cancer cells.[58] In response to integrin activation, FAK or the related kinase RAFTK recruits NEDD9 into a focal adhesion site, binds it via the N-terminal SH3 domain and phosphorylates the NEDD9 Src-binding site. This allows SRC or SRC family kinase to bind NEDD9 via its SH2 domain. Phosphorylation of the NEDD9 substrate domain by Src and other kinases results in the creation of binding sites for Crk and other adaptors that associate with SH2 binding motifs. NEDD9 Crk complexes activate Rho and Ras family GTPases via the recruitment of their nucleotide exchange factors (GEFs), such as DOCK1, DOCK3 [36] DOCK180 and C3G.[59]
These GTPases regulate cell motility, proliferation and also contribute to tumor progression and invasion. In many cell types, NEDD9 overexpression increases spreading and crescent morphology (an indicator of high motility).[54] However, in fibroblasts, some work has found that absence of NEDD9 leads to more rapid focal adhesion turnover, which led to increase of migration in NEDD9-/- compared to wild type.[58]
In cancer cells, NEDD9 can drive mesenchymal-type movement by activating RAC1 GTPase and WAVE in complex with its GEF DOCK3, which in turn cause inhibition of GTPase Rho and amoeboid movement.[60] Invasion is accompanied by proteolysis of the ECM through activation of MMP14, MMP2 and MMP9 metalloproteinases.[61]
Chemokine receptors, TCR, BCR/ABL, Fyn, Lck kinases
NEDD9 is involved in chemokine-induced T cell migration and T cell receptor (TCR)–mediated integrin activation. In lymphocytes, integrin or TCR signaling induces NEDD9 phosphorylation by tyrosine kinases Fyn and Lck (SRC family kinases), which is essential for T cell migration.[62] In addition, in response to chemokine signals, Abl family kinases promote GTPase RAP1 activation by phosphorylating of NEDD9;[63] NEDD9 associates with the transducer protein Chat-H/SHEP1/NSP3, a member of the NSP protein family, further supporting RAP1 activation, cell migration, and adhesion.[64] In B cells, NEDD9 association with NSP3 enhances integrin-mediated NEDD9 serine/threonine hyperphosphorylation following B cell receptor (BCR) ligation, promoting B lymphocyte adhesion, motility and homing into marginal zones of spleen [65] Estrogen Receptor. The NEDD9 interactors p130/CAS and the NSP protein NSP2/BCAR3 are implicated in antiestrogen resistance [66][67] and breast cancer progression [68] Some data suggests a role for NEDD9 in the cellular response to estrogen, including the progression to anti-estrogen resistance, breast cancer progression and invasion [69][70][71]
RTKs (EGFR). NEDD9 also contributes to the transduction of signals downstream receptor tyrosine kinases (RTKs). A role for NEDD9 in signaling crosstalk between epidermal growth factor receptor (EGFR) and integrins was established in non-small lung cancer (NSLC). It was shown that inhibition of EGFR reduces the tyrosine phosphorylation of NEDD9.[72] Nedd9 interacts directly with the EGFR effector protein Shc, positioning it to affect downstream signaling relevant to EGFR; mice lacking Nedd9 have depressed activity of the EGFR effectors ERK and AKT.[73] NSP proteins are also multidomain scaffolds, which bind activated RTKs in response to extracellular stimuli and recruit both NEDD9 and BCAR1 to assist in integrating signaling between RTKs and integrins. NEDD9 is also activated by PDGF [74] and other RTKs, although more study is required.
TGF-beta
TGF-beta is a regulator of tissue remodeling and epithelial-mesenchymal transition (EMT) in development, and promotes metastasis in cancer. A number of studies have identified NEDD9 as a downstream effector in the TGF-beta signaling pathway, essential for promoting EMT.[21][55][75][76][77] In MCF-7 cells, NEDD9 negatively regulates expression of the epithelial protein E-cadherin, preventing association of E-cadherin with cell membrane and activating SRC-kinase.[78] Activated SRC provides internalization and lysosomal degradation of E-cadherin.[78] Consistent with these findings is a study demonstrating downregulation of epithelial markers (E-cadherin, occludin, β-catenin) and concurrent upregulation of mesenchymal markers (N-cadherin, vimentin, fibronectin) in response to NEDD9 overexpression in MCF-10 cells.[79]
Aurora-A
NEDD9 binds directly to the Aurora-A mitotic kinase at the centrosome, and promotes its activity, allowing cells to enter mitosis.[43][80] Degradation of NEDD9 at the end of mitosis contributes to timely Aurora-A degradation.[43][80][81] Cells overexpressing NEDD9 exhibit deficient cytokinesis resulting in the accumulation of multipolar mitotic spindles and abnormal numbers of centrosomes. On the other hand, cells with depleted NEDD9 have prematurely separated centrosomes and are deficient in microtubule organizing activity during mitosis, leading to an abundance of monopolar or asymmetric spindles,[43] preventing cells from entering mitosis. NEDD9 also regulates Aurora-A activation at the basal body of cilia as cells resorb cilia during early G1.[57] Cilia are small organelles that protrude from the surface of adherent cells that are the obligate site of action for proteins such as Hedgehog, and the polycystins: by influencing ciliary stability, NEDD9 is positioned to affect these signaling systems. Interaction of NEDD9 with Aurora A kinase may also play a role in tumor invasion. NEDD9 binds to and regulates acetylation of cortactin (CTTN) in an Aurora A kinase (AURKA)/HDAC6–dependent manner. The knockdown of NEDD9 or AURKA results in an increase in the amount of acetylated CTTN and a decrease in the binding of CTTN to F-actin. Overexpression of the deacetylation mimicking (9KR) mutant of CTTN is sufficient to restore actin dynamics at the leading edge and migration proficiency of the tumor cells. Inhibition of AURKA and HDAC6 activity by alisertib and tubastatin A in xenograft models of breast cancer has led to a decrease in the number of pulmonary metastases.[82]
Clinical significance
Transgenic mice with homozygous depletion of NEDD9 are vital and fertile, but have immunological abnormalities that result in pre-malignant conditions later in life, defects are initially subtle, but increase in later life; B cell homing to the spleen and lymphocyte trafficking are deficient.[73][83]
Alzheimer's disease
The NEDD9 rs760678 SNP located in an intronic region, has been studied for a possible association with late onset Alzheimer's disease (LOAD).[84][85][86][87][88] However, in 2012, Wang et al., performed a meta-analysis and concluded that more studies are required for solid conclusions.[87] This SNP and relevant signaling is discussed more fully in.[89]
Cancer
Altered (typically elevated) expression of NEDD9 is strongly associated with cancer. NEDD9 is rarely if ever mutated, but frequently show altered expression or phosphorylation (associated with increased activity) in pathological conditions including immune cell dysfunction and cancer. NEDD9 overexpression is documented to occur and in some cases linked the process of tumorigenesis of many different malignances. Besides examples in breast cancer discussed above, these malignancies include colon,[14][15][18][90] pancreatic,[91] head and neck,[92] ovarian,[93] gastric,[94] lung,[95] genitourinary (including prostate),[23][96] liver,[19] and kidney cancer,[17][24] gastrointestinal stromal tumors,[97] glioblastoma,[22][74][98] and neuroblastoma.[9][10][58]
Other disease
Nedd9 expression may be important for recovery from stroke. Nedd9 is upregulated in the neurons of the cerebral cortex and hippocampus after transient global ischemia in rats. Induced Nedd9 is tyrosine phosphorylated, bound to FAK in dendrite and soma of neurons, and promotes neurite outgrowth, contributing into recovery of neurologic function after cerebral ischemia.[99] Nedd9 has recently been implicated in the pathogenesis of autosomal dominant polycystic kidney disease (ADPKD). NEDD9 expression is elevated in human autosomal dominant polycystic kidney disease (ADPKD) and in mouse ADPKD models, and ADPKD-prone mice lacking NEDD9 developed a more severe form of ADPKD than those with normal NEDD9.[100]
Therapeutic potential
Because of its roles in cancer, several studies have considered the potential value of NEDD9 as a therapeutic target or therapeutic guide. Because of lack of a kinase domain, or any defined catalytic domain, and because it is entirely intracellular, NEDD9 is a difficult molecule to target. Because NEDD9 serves as a scaffolding molecule for other signaling proteins that play significant roles in cancer development, the effects of NEDD9 overexpression in supporting metastasis could in theory be mitigated by inhibition of its downstream targets. In one study, deletion of Nedd9 in MMTV-neu mammary tumors increased their sensitivity to inhbitiors of FAK and SRC.[101] NEDD9 depletion sensitizes breast tumor cell lines to the Aurora A inhibitor alisertib.[81] Consideration of NEDD9 as a biomarker for therapeutic response is a promising research direction.
Interactions
NEDD9 has been shown to interact with:
Notes
References
- GRCh38: Ensembl release 89: ENSG00000111859 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000021365 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: NEDD9 neural precursor cell expressed, developmentally down-regulated 9".
- Kumar S, Tomooka Y, Noda M (1992). "Identification of a set of genes with developmentally down-regulated expression in the mouse brain". Biochem. Biophys. Res. Commun. 185 (3): 1155–61. doi:10.1016/0006-291x(92)91747-e. PMID 1378265.
- Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA (1996). "Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae". Mol. Cell. Biol. 16 (7): 3327–37. doi:10.1128/mcb.16.7.3327. PMC 231327. PMID 8668148.
- Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C (1996). "Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes". J. Exp. Med. 184 (4): 1365–75. doi:10.1084/jem.184.4.1365. PMC 2192828. PMID 8879209.
- Merrill RA, Ahrens JM, Kaiser ME, Federhart KS, Poon VY, Clagett-Dame M (2004). "All-trans retinoic acid-responsive genes identified in the human SH-SY5Y neuroblastoma cell line and their regulated expression in the nervous system of early embryos". Biol. Chem. 385 (7): 605–14. doi:10.1515/BC.2004.075. PMID 15318809.
- Merrill RA, See AW, Wertheim ML, Clagett-Dame M (2004). "Crk-associated substrate (Cas) family member, NEDD9, is regulated in human neuroblastoma cells and in the embryonic hindbrain by all-trans retinoic acid". Dev. Dyn. 231 (3): 564–75. doi:10.1002/dvdy.20159. PMID 15376324.
- Knutson DC, Clagett-Dame M (2015). "A complex RARE is required for the majority of Nedd9 embryonic expression". Transgenic Res. 24 (1): 123–34. doi:10.1007/s11248-014-9825-9. PMC 4274375. PMID 25120220.
- Bui LC, Tomkiewicz C, Chevallier A, Pierre S, Bats AS, Mota S, Raingeaud J, Pierre J, Diry M, Transy C, Garlatti M, Barouki R, Coumoul X (2009). "Nedd9/Hef1/Cas-L mediates the effects of environmental pollutants on cell migration and plasticity". Oncogene. 28 (41): 3642–51. doi:10.1038/onc.2009.224. PMID 19648964.
- Hammerich-Hille S, Kaipparettu BA, Tsimelzon A, Creighton CJ, Jiang S, Polo JM, Melnick A, Meyer R, Oesterreich S (2010). "SAFB1 mediates repression of immune regulators and apoptotic genes in breast cancer cells". J. Biol. Chem. 285 (6): 3608–16. doi:10.1074/jbc.M109.066431. PMC 2823501. PMID 19901029.
- Li Y, Bavarva JH, Wang Z, Guo J, Qian C, Thibodeau SN, Golemis EA, Liu W (2011). "HEF1, a novel target of Wnt signaling, promotes colonic cell migration and cancer progression". Oncogene. 30 (23): 2633–43. doi:10.1038/onc.2010.632. PMC 3164309. PMID 21317929.
- Kim SH, Xia D, Kim SW, Holla V, Menter DG, Dubois RN (2010). "Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells". Cancer Res. 70 (10): 4054–63. doi:10.1158/0008-5472.CAN-09-2110. PMC 2871069. PMID 20442290.
- Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, Roubelakis M, Sweeney D, Moukayed M, Harris AL, Davies K, Watt SM (2007). "Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia". Stem Cells. 25 (4): 1003–12. doi:10.1634/stemcells.2006-0398. PMID 17185612.
- Xu J, Li H, Wang B, Xu Y, Yang J, Zhang X, Harten SK, Shukla D, Maxwell PH, Pei D, Esteban MA (2010). "VHL inactivation induces HEF1 and Aurora kinase A". J. Am. Soc. Nephrol. 21 (12): 2041–6. doi:10.1681/ASN.2010040345. PMC 3014016. PMID 20864688.
- Xia D, Holla VR, Wang D, Menter DG, DuBois RN (2010). "HEF1 is a crucial mediator of the proliferative effects of prostaglandin E(2) on colon cancer cells". Cancer Res. 70 (2): 824–31. doi:10.1158/0008-5472.CAN-09-2105. PMC 2943830. PMID 20068165.
- Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, Zhang Y, Hu H, Fan D, Nie Y, Wu K (2013). "Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma". Hepatology. 57 (2): 610–24. doi:10.1002/hep.26029. PMID 22911555.
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M (2011). "The transcription factor PAX5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells". EMBO J. 30 (12): 2388–404. doi:10.1038/emboj.2011.140. PMC 3116275. PMID 21552207.
- Zheng M, McKeown-Longo PJ (2002). "Regulation of HEF1 expression and phosphorylation by TGF-beta 1 and cell adhesion". J. Biol. Chem. 277 (42): 39599–608. doi:10.1074/jbc.M202263200. PMID 12189134.
- Speranza MC, Frattini V, Pisati F, Kapetis D, Porrati P, Eoli M, Pellegatta S, Finocchiaro G (2012). "NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma". Oncotarget. 3 (7): 723–34. doi:10.18632/oncotarget.547. PMC 3443255. PMID 22869051.
- Guo W, Ren D, Chen X, Tu X, Huang S, Wang M, Song L, Zou X, Peng X (2013). "HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145". J. Cell. Biochem. 114 (7): 1606–15. doi:10.1002/jcb.24502. PMID 23355420.
- Lu R, Ji Z, Li X, Zhai Q, Zhao C, Jiang Z, Zhang S, Nie L, Yu Z (2014). "miR-145 functions as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma". J. Cancer Res. Clin. Oncol. 140 (3): 387–97. doi:10.1007/s00432-013-1577-z. PMID 24384875.
- Malleter M, Jacquot C, Moreau D, Tomasoni C, Tsvetanova M, Chinou I, Juge M, Pineau A, Le Pape P, Roussakis C (2010). "A novel large regulator RNA, B2, partially overlaps the HEF1/NEDD9/Cas-L gene". Int. J. Mol. Med. 25 (6): 897–903. doi:10.3892/ijmm_00000420. PMID 20428794.
- Aquino JB, Marmigère F, Lallemend F, Lundgren TK, Villar MJ, Wegner M, Ernfors P (2008). "Differential expression and dynamic changes of murine NEDD9 in progenitor cells of diverse tissues". Gene Expr. Patterns. 8 (4): 217–26. doi:10.1016/j.gep.2008.01.001. PMID 18282814.
- Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H (1994). "A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner". EMBO J. 13 (16): 3748–56. doi:10.1002/j.1460-2075.1994.tb06684.x. PMC 395286. PMID 8070403.
- Ishino M, Ohba T, Sasaki H, Sasaki T (1995). "Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which contains a Src homology 3 domain and associates with Fyn". Oncogene. 11 (11): 2331–8. PMID 8570184.
- Alexandropoulos K, Cheng G, Baltimore D (1995). "Proline-rich sequences that bind to Src homology 3 domains with individual specificities". Proc. Natl. Acad. Sci. U.S.A. 92 (8): 3110–4. Bibcode:1995PNAS...92.3110A. doi:10.1073/pnas.92.8.3110. PMC 42114. PMID 7536925.
- Singh MK, Dadke D, Nicolas E, Serebriiskii IG, Apostolou S, Canutescu A, Egleston BL, Golemis EA (2008). "A novel Cas family member, HEPL, regulates FAK and cell spreading". Mol. Biol. Cell. 19 (4): 1627–36. doi:10.1091/mbc.E07-09-0953. PMC 2291417. PMID 18256281.
- Huang Z, Yazdani U, Thompson-Peer KL, Kolodkin AL, Terman JR (2007). "Crk-associated substrate (Cas) signaling protein functions with integrins to specify axon guidance during development". Development. 134 (12): 2337–47. doi:10.1242/dev.004242. PMID 17537798.
- Tikhmyanova N, Tulin AV, Roegiers F, Golemis EA (2010). "Dcas supports cell polarization and cell-cell adhesion complexes in development". PLOS ONE. 5 (8): e12369. Bibcode:2010PLoSO...512369T. doi:10.1371/journal.pone.0012369. PMC 2927436. PMID 20808771.
- Nikonova AS, Gaponova AV, Kudinov AE, Golemis EA (2014). "CAS proteins in health and disease: an update". IUBMB Life. 66 (6): 387–95. doi:10.1002/iub.1282. PMC 4111207. PMID 24962474.
- Singh M, Cowell L, Seo S, O'Neill G, Golemis E (2007). "Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle". Cell Biochem. Biophys. 48 (1): 54–72. doi:10.1007/s12013-007-0036-3. PMC 1976382. PMID 17703068.
- Tikhmyanova N, Little JL, Golemis EA (2010). "CAS proteins in normal and pathological cell growth control". Cell. Mol. Life Sci. 67 (7): 1025–48. doi:10.1007/s00018-009-0213-1. PMC 2836406. PMID 19937461.
- Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P (2010). "Integrin signalling adaptors: not only figurants in the cancer story". Nat. Rev. Cancer. 10 (12): 858–70. doi:10.1038/nrc2967. hdl:2318/80156. PMID 21102636.
- O'Neill GM, Fashena SJ, Golemis EA (2000). "Integrin signalling: a new Cas(t) of characters enters the stage". Trends Cell Biol. 10 (3): 111–9. doi:10.1016/s0962-8924(99)01714-6. PMID 10675905.
- Kirsch KH, Georgescu MM, Hanafusa H (1998). "Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G". J. Biol. Chem. 273 (40): 25673–9. doi:10.1074/jbc.273.40.25673. PMID 9748234.
- Garton AJ, Burnham MR, Bouton AH, Tonks NK (1997). "Association of PTP-PEST with the SH3 domain of p130cas; a novel mechanism of protein tyrosine phosphatase substrate recognition". Oncogene. 15 (8): 877–85. doi:10.1038/sj.onc.1201279. PMID 9285683.
- Liu F, Hill DE, Chernoff J (1996). "Direct binding of the proline-rich region of protein tyrosine phosphatase 1B to the Src homology 3 domain of p130(Cas)". J. Biol. Chem. 271 (49): 31290–5. doi:10.1074/jbc.271.49.31290. PMID 8940134.
- Nakamoto T, Yamagata T, Sakai R, Ogawa S, Honda H, Ueno H, Hirano N, Yazaki Y, Hirai H (2000). "CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases". Mol. Cell. Biol. 20 (5): 1649–58. doi:10.1128/mcb.20.5.1649-1658.2000. PMC 85348. PMID 10669742.
- Tamada M, Sheetz MP, Sawada Y (2004). "Activation of a signaling cascade by cytoskeleton stretch". Dev. Cell. 7 (5): 709–18. doi:10.1016/j.devcel.2004.08.021. PMID 15525532.
- Pugacheva EN, Golemis EA (2005). "The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome". Nat. Cell Biol. 7 (10): 937–46. doi:10.1038/ncb1309. PMC 2652766. PMID 16184168.
- Briknarová K, Nasertorabi F, Havert ML, Eggleston E, Hoyt DW, Li C, Olson AJ, Vuori K, Ely KR (2005). "The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle". J. Biol. Chem. 280 (23): 21908–14. doi:10.1074/jbc.M501258200. PMID 15795225.
- Arold ST, Hoellerer MK, Noble ME (2002). "The structural basis of localization and signaling by the focal adhesion targeting domain". Structure. 10 (3): 319–27. doi:10.1016/s0969-2126(02)00717-7. PMID 12005431.
- Hayashi I, Vuori K, Liddington RC (2002). "The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin". Nat. Struct. Biol. 9 (2): 101–6. doi:10.1038/nsb755. PMID 11799401.
- Wallez Y, Mace PD, Pasquale EB, Riedl SJ (2012). "NSP-CAS Protein Complexes: Emerging Signaling Modules in Cancer". Genes Cancer. 3 (5–6): 382–93. doi:10.1177/1947601912460050. PMC 3513790. PMID 23226576.
- Mace PD, Wallez Y, Dobaczewska MK, Lee JJ, Robinson H, Pasquale EB, Riedl SJ (2011). "NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling". Nat. Struct. Mol. Biol. 18 (12): 1381–7. doi:10.1038/nsmb.2152. PMC 3230775. PMID 22081014.
- Law SF, Zhang YZ, Fashena SJ, Toby G, Estojak J, Golemis EA (1999). "Dimerization of the docking/adaptor protein HEF1 via a carboxy-terminal helix-loop-helix domain". Exp. Cell Res. 252 (1): 224–35. doi:10.1006/excr.1999.4609. PMID 10502414.
- Law SF, Zhang YZ, Klein-Szanto AJ, Golemis EA (1998). "Cell cycle-regulated processing of HEF1 to multiple protein forms differentially targeted to multiple subcellular compartments". Mol. Cell. Biol. 18 (6): 3540–51. doi:10.1128/MCB.18.6.3540. PMC 108935. PMID 9584194.
- Zheng M, McKeown-Longo PJ (2006). "Cell adhesion regulates Ser/Thr phosphorylation and proteasomal degradation of HEF1". J. Cell Sci. 119 (Pt 1): 96–103. doi:10.1242/jcs.02712. PMID 16352661.
- Bradbury P, Mahmassani M, Zhong J, Turner K, Paul A, Verrills NM, O'Neill GM (2012). "PP2A phosphatase suppresses function of the mesenchymal invasion regulator NEDD9". Biochim. Biophys. Acta. 1823 (2): 290–7. doi:10.1016/j.bbamcr.2011.10.011. PMID 22061964.
- Law SF, O'Neill GM, Fashena SJ, Einarson MB, Golemis EA (2000). "The docking protein HEF1 is an apoptotic mediator at focal adhesion sites". Mol. Cell. Biol. 20 (14): 5184–95. doi:10.1128/mcb.20.14.5184-5195.2000. PMC 85967. PMID 10866674.
- O'Neill GM, Golemis EA (2001). "Proteolysis of the docking protein HEF1 and implications for focal adhesion dynamics". Mol. Cell. Biol. 21 (15): 5094–108. doi:10.1128/MCB.21.15.5094-5108.2001. PMC 87235. PMID 11438665.
- Liu X, Elia AE, Law SF, Golemis EA, Farley J, Wang T (2000). "A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member HEF1". EMBO J. 19 (24): 6759–69. doi:10.1093/emboj/19.24.6759. PMC 305889. PMID 11118211.
- Nourry C, Maksumova L, Pang M, Liu X, Wang T (2004). "Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1". BMC Cell Biol. 5: 20. doi:10.1186/1471-2121-5-20. PMC 420458. PMID 15144564.
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA (2007). "HEF1-dependent Aurora A activation induces disassembly of the primary cilium". Cell. 129 (7): 1351–63. doi:10.1016/j.cell.2007.04.035. PMC 2504417. PMID 17604723.
- Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, Fabry B, O'Neill GM (2012). "NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration". PLOS ONE. 7 (4): e35058. Bibcode:2012PLoSO...735058Z. doi:10.1371/journal.pone.0035058. PMC 3324407. PMID 22509381.
- Guerrero MS, Parsons JT, Bouton AH (2012). "Cas and NEDD9 Contribute to Tumor Progression through Dynamic Regulation of the Cytoskeleton". Genes Cancer. 3 (5–6): 371–81. doi:10.1177/1947601912458585. PMC 3513795. PMID 23226575.
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ (2008). "Rac activation and inactivation control plasticity of tumor cell movement". Cell. 135 (3): 510–23. doi:10.1016/j.cell.2008.09.043. PMID 18984162.
- McLaughlin SL, Ice RJ, Rajulapati A, Kozyulina PY, Livengood RH, Kozyreva VK, Loskutov YV, Culp MV, Weed SA, Ivanov AV, Pugacheva EN (2014). "NEDD9 depletion leads to MMP14 inactivation by TIMP2 and prevents invasion and metastasis". Mol. Cancer Res. 12 (1): 69–81. doi:10.1158/1541-7786.MCR-13-0300. PMC 3946989. PMID 24202705.
- Kanda H, Mimura T, Hamasaki K, Yamamoto K, Yazaki Y, Hirai H, Nojima Y (1999). "Fyn and Lck tyrosine kinases regulate tyrosine phosphorylation of p105CasL, a member of the p130Cas docking protein family, in T-cell receptor-mediated signalling". Immunology. 97 (1): 56–61. doi:10.1046/j.1365-2567.1999.00753.x. PMC 2326814. PMID 10447714.
- Malherbe LP, Wang D (2012). "Tyrosine kinases EnAbling adaptor molecules for chemokine-induced Rap1 activation in T cells". Sci Signal. 5 (235): pe33. doi:10.1126/scisignal.2003383. PMC 4307919. PMID 22855504.
- Regelmann AG, Danzl NM, Wanjalla C, Alexandropoulos K (2006). "The hematopoietic isoform of Cas-Hef1-associated signal transducer regulates chemokine-induced inside-out signaling and T cell trafficking". Immunity. 25 (6): 907–18. doi:10.1016/j.immuni.2006.09.014. PMID 17174122.
- Browne CD, Hoefer MM, Chintalapati SK, Cato MH, Wallez Y, Ostertag DV, Pasquale EB, Rickert RC (2010). "SHEP1 partners with CasL to promote marginal zone B-cell maturation". Proc. Natl. Acad. Sci. U.S.A. 107 (44): 18944–9. Bibcode:2010PNAS..10718944B. doi:10.1073/pnas.1007558107. PMC 2973925. PMID 20956287.
- Wallez Y, Riedl SJ, Pasquale EB (2014). "Association of the breast cancer antiestrogen resistance protein 1 (BCAR1) and BCAR3 scaffolding proteins in cell signaling and antiestrogen resistance". J. Biol. Chem. 289 (15): 10431–44. doi:10.1074/jbc.M113.541839. PMC 4036165. PMID 24584939.
- Brinkman A, van der Flier S, Kok EM, Dorssers LC (2000). "BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells". J. Natl. Cancer Inst. 92 (2): 112–20. doi:10.1093/jnci/92.2.112. PMID 10639512.
- Arpaia E, Blaser H, Quintela-Fandino M, Duncan G, Leong HS, Ablack A, Nambiar SC, Lind EF, Silvester J, Fleming CK, Rufini A, Tusche MW, Brüstle A, Ohashi PS, Lewis JD, Mak TW (2012). "The interaction between caveolin-1 and Rho-GTPases promotes metastasis by controlling the expression of alpha5-integrin and the activation of Src, Ras and Erk". Oncogene. 31 (7): 884–96. doi:10.1038/onc.2011.288. PMC 3289793. PMID 21765460.
- van Agthoven T, Godinho MF, Wulfkuhle JD, Petricoin EF, Dorssers LC (2012). "Protein pathway activation mapping reveals molecular networks associated with antiestrogen resistance in breast cancer cell lines". Int. J. Cancer. 131 (9): 1998–2007. doi:10.1002/ijc.27489. PMID 22328489.
- Garron ML, Arsenieva D, Zhong J, Bloom AB, Lerner A, O'Neill GM, Arold ST (2009). "Structural insights into the association between BCAR3 and Cas family members, an atypical complex implicated in anti-oestrogen resistance". J. Mol. Biol. 386 (1): 190–203. doi:10.1016/j.jmb.2008.12.010. PMID 19103205.
- Bradshaw LN, Zhong J, Bradbury P, Mahmassani M, Smith JL, Ammit AJ, O'Neill GM (2011). "Estradiol stabilizes the 105-kDa phospho-form of the adhesion docking protein NEDD9 and suppresses NEDD9-dependent cell spreading in breast cancer cells". Biochim. Biophys. Acta. 1813 (2): 340–5. doi:10.1016/j.bbamcr.2010.11.018. PMID 21145356.
- Kondo S, Iwata S, Yamada T, Inoue Y, Ichihara H, Kichikawa Y, Katayose T, Souta-Kuribara A, Yamazaki H, Hosono O, Kawasaki H, Tanaka H, Hayashi Y, Sakamoto M, Kamiya K, Dang NH, Morimoto C (2012). "Impact of the integrin signaling adaptor protein NEDD9 on prognosis and metastatic behavior of human lung cancer". Clin. Cancer Res. 18 (22): 6326–38. doi:10.1158/1078-0432.CCR-11-2162. PMID 23037767.
- Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, Seo S, Kurokawa M, Egleston BL, Klein-Szanto A, Pugacheva EN, Hardy RR, Wolfson M, Connolly DC, Golemis EA (2009). "NEDD9 promotes oncogenic signaling in mammary tumor development". Cancer Res. 69 (18): 7198–206. doi:10.1158/0008-5472.CAN-09-0795. PMC 2758619. PMID 19738060.
- Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, Wang W, Grammer JR, Gladson CL (2006). "HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells". Oncogene. 25 (12): 1721–32. doi:10.1038/sj.onc.1209199. PMID 16288224.
- Inamoto S, Iwata S, Inamoto T, Nomura S, Sasaki T, Urasaki Y, Hosono O, Kawasaki H, Tanaka H, Dang NH, Morimoto C (2007). "Crk-associated substrate lymphocyte type regulates transforming growth factor-beta signaling by inhibiting Smad6 and Smad7". Oncogene. 26 (6): 893–904. doi:10.1038/sj.onc.1209848. PMID 16909115.
- Vogel T, Ahrens S, Büttner N, Krieglstein K (2010). "Transforming growth factor beta promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component". Cereb. Cortex. 20 (3): 661–71. doi:10.1093/cercor/bhp134. PMC 2820705. PMID 19587023.
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E (2009). "Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility". Nat. Cell Biol. 11 (11): 1287–96. doi:10.1038/ncb1973. PMC 2773241. PMID 19838175.
- Tikhmyanova N, Golemis EA (2011). "NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation". PLOS ONE. 6 (7): e22102. Bibcode:2011PLoSO...622102T. doi:10.1371/journal.pone.0022102. PMC 3134485. PMID 21765937.
- Kong C, Wang C, Wang L, Ma M, Niu C, Sun X, Du J, Dong Z, Zhu S, Lu J, Huang B (2011). "NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer". PLOS ONE. 6 (7): e22666. Bibcode:2011PLoSO...622666K. doi:10.1371/journal.pone.0022666. PMC 3145662. PMID 21829474.
- Pugacheva EN, Golemis EA (2006). "HEF1-aurora A interactions: points of dialog between the cell cycle and cell attachment signaling networks". Cell Cycle. 5 (4): 384–91. doi:10.4161/cc.5.4.2439. PMC 2547350. PMID 16479169.
- Ice RJ, McLaughlin SL, Livengood RH, Culp MV, Eddy ER, Ivanov AV, Pugacheva EN (2013). "NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors". Cancer Res. 73 (10): 3168–80. doi:10.1158/0008-5472.CAN-12-4008. PMC 3667743. PMID 23539442.
- Kozyreva VK, McLaughlin SL, Livengood RH, Calkins RA, Kelley LC, Rajulapati A, Ice RJ, Smolkin MB, Weed SA, Pugacheva EN (2014). "NEDD9 regulates actin dynamics through cortactin deacetylation in an AURKA/HDAC6-dependent manner". Mol. Cancer Res. 12 (5): 681–93. doi:10.1158/1541-7786.MCR-13-0654. PMC 4020952. PMID 24574519.
- Seo S, Asai T, Saito T, Suzuki T, Morishita Y, Nakamoto T, Ichikawa M, Yamamoto G, Kawazu M, Yamagata T, Sakai R, Mitani K, Ogawa S, Kurokawa M, Chiba S, Hirai H (2005). "Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance". J. Immunol. 175 (6): 3492–501. doi:10.4049/jimmunol.175.6.3492. PMID 16148091.
- Chapuis J, Moisan F, Mellick G, Elbaz A, Silburn P, Pasquier F, Hannequin D, Lendon C, Campion D, Amouyel P, Lambert JC (2008). "Association study of the NEDD9 gene with the risk of developing Alzheimer's and Parkinson's disease". Hum. Mol. Genet. 17 (18): 2863–7. CiteSeerX 10.1.1.1026.7862. doi:10.1093/hmg/ddn183. PMID 18579580.
- Li Y, Grupe A, Rowland C, Holmans P, Segurado R, Abraham R, Jones L, Catanese J, Ross D, Mayo K, Martinez M, Hollingworth P, Goate A, Cairns NJ, Racette BA, Perlmutter JS, O'Donovan MC, Morris JC, Brayne C, Rubinsztein DC, Lovestone S, Thal LJ, Owen MJ, Williams J (2008). "Evidence that common variation in NEDD9 is associated with susceptibility to late-onset Alzheimer's and Parkinson's disease". Hum. Mol. Genet. 17 (5): 759–67. doi:10.1093/hmg/ddm348. PMID 18063669.
- Tedde A, Bagnoli S, Piaceri I, Lucenteforte E, Bessi V, Bracco L, Mugelli A, Sorbi S, Nacmias B (2010). "Different implication of NEDD9 genetic variant in early and late-onset Alzheimer's disease". Neurosci. Lett. 477 (3): 121–3. doi:10.1016/j.neulet.2010.04.046. PMID 20430066.
- Wang Y, Bi L, Wang H, Li Y, Di Q, Xu W, Qian Y (2012). "NEDD9 rs760678 polymorphism and the risk of Alzheimer's disease: a meta-analysis". Neurosci. Lett. 527 (2): 121–5. doi:10.1016/j.neulet.2012.08.044. PMID 22963925.
- Xing YY, Yu JT, Yan WJ, Chen W, Zhong XL, Jiang H, Wang P, Tan L (2011). "NEDD9 is genetically associated with Alzheimer's disease in a Han Chinese population". Brain Res. 1369: 230–4. doi:10.1016/j.brainres.2010.10.113. PMID 21059344.
- Beck, T.N.; et al. "Adaptors for disorders of the brain? The cancer signaling proteins NEDD9, CASS4, and PTK2B in Alzheimer's disease" (Oncoscience, 2014. 1(7): p. 486–503). Cite journal requires
|journal=(help) - Li P, Zhou H, Zhu X, Ma G, Liu C, Lin B, Mao W (2014). "High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients". Int J Clin Exp Pathol. 7 (5): 2565–70. PMC 4069898. PMID 24966970.
- Xue YZ, Sheng YY, Liu ZL, Wei ZQ, Cao HY, Wu YM, Lu YF, Yu LH, Li JP, Li ZS (2013). "Expression of NEDD9 in pancreatic ductal adenocarcinoma and its clinical significance". Tumour Biol. 34 (2): 895–9. doi:10.1007/s13277-012-0624-8. PMID 23247867.
- Lucas JT, Salimath BP, Slomiany MG, Rosenzweig SA (2010). "Regulation of invasive behavior by vascular endothelial growth factor is HEF1-dependent". Oncogene. 29 (31): 4449–59. doi:10.1038/onc.2010.185. PMC 2921319. PMID 20498643.
- Wang H, Mu X, Zhou S, Zhang J, Dai J, Tang L, Xiao L, Duan Z, Jia L, Chen S (2014). "NEDD9 overexpression is associated with the progression of and an unfavorable prognosis in epithelial ovarian cancer". Hum. Pathol. 45 (2): 401–8. doi:10.1016/j.humpath.2013.10.005. PMID 24439227.
- Zhang Q, Wang H, Ma Y, Zhang J, He X, Ma J, Zhao ZS (2014). "Overexpression of Nedd9 is a prognostic marker of human gastric cancer". Med. Oncol. 31 (7): 33. doi:10.1007/s12032-014-0033-5. PMID 24906654.
- Jin Y, Li F, Zheng C, Wang Y, Fang Z, Guo C, Wang X, Liu H, Deng L, Li C, Wang H, Chen H, Feng Y, Ji H (2014). "NEDD9 promotes lung cancer metastasis through epithelial-mesenchymal transition". Int. J. Cancer. 134 (10): 2294–304. doi:10.1002/ijc.28568. PMID 24174333.
- Morimoto K, Tanaka T, Nitta Y, Ohnishi K, Kawashima H, Nakatani T (2014). "NEDD9 crucially regulates TGF-β-triggered epithelial-mesenchymal transition and cell invasion in prostate cancer cells: involvement in cancer progressiveness". Prostate. 74 (8): 901–10. doi:10.1002/pros.22809. PMID 24728978.
- Thao le B, Vu HA, Yasuda K, Taniguchi S, Yagasaki F, Taguchi T, Watanabe T, Sato Y (2009). "Cas-L was overexpressed in imatinib-resistant gastrointestinal stromal tumor cells". Cancer Biol. Ther. 8 (8): 683–8. doi:10.4161/cbt.8.8.7779. PMID 19417561.
- Ismail HM (2012). "Overexpression of s6 kinase 1 in brain tumours is associated with induction of hypoxia-responsive genes and predicts patients' survival". J Oncol. 2012: 1–10. doi:10.1155/2012/416927. PMC 3335255. PMID 22570651.
- Sasaki T, Iwata S, Okano HJ, Urasaki Y, Hamada J, Tanaka H, Dang NH, Okano H, Morimoto C (2005). "Nedd9 protein, a Cas-L homologue, is upregulated after transient global ischemia in rats: possible involvement of Nedd9 in the differentiation of neurons after ischemia". Stroke. 36 (11): 2457–62. doi:10.1161/01.STR.0000185672.10390.30. PMID 16210561.
- Nikonova AS, Plotnikova OV, Serzhanova V, Efimov A, Bogush I, Cai KQ, Hensley HH, Egleston BL, Klein-Szanto A, Seeger-Nukpezah T, Golemis EA (2014). "Nedd9 restrains renal cystogenesis in Pkd1-/- mice". Proc. Natl. Acad. Sci. U.S.A. 111 (35): 12859–64. Bibcode:2014PNAS..11112859N. doi:10.1073/pnas.1405362111. PMC 4156736. PMID 25139996.
- Little JL, Serzhanova V, Izumchenko E, Egleston BL, Parise E, Klein-Szanto AJ, Loudon G, Shubina M, Seo S, Kurokawa M, Ochs MF, Golemis EA (2014). "A requirement for Nedd9 in luminal progenitor cells prior to mammary tumorigenesis in MMTV-HER2/ErbB2 mice". Oncogene. 33 (4): 411–20. doi:10.1038/onc.2012.607. PMC 3628996. PMID 23318423.
- Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C (October 1996). "Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes". J. Exp. Med. 184 (4): 1365–75. doi:10.1084/jem.184.4.1365. PMC 2192828. PMID 8879209.
- Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA (July 1996). "Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae". Mol. Cell. Biol. 16 (7): 3327–37. doi:10.1128/mcb.16.7.3327. PMC 231327. PMID 8668148.
- Nourry C, Maksumova L, Pang M, Liu X, Wang T (May 2004). "Direct interaction between Smad3, APC10, CDH1 and HEF1 in proteasomal degradation of HEF1". BMC Cell Biol. 5: 20. doi:10.1186/1471-2121-5-20. PMC 420458. PMID 15144564.
- Ohashi Y, Tachibana K, Kamiguchi K, Fujita H, Morimoto C (March 1998). "T cell receptor-mediated tyrosine phosphorylation of Cas-L, a 105-kDa Crk-associated substrate-related protein, and its association of Crk and C3G". J. Biol. Chem. 273 (11): 6446–51. doi:10.1074/jbc.273.11.6446. PMID 9497377.
- Manié SN, Beck AR, Astier A, Law SF, Canty T, Hirai H, Druker BJ, Avraham H, Haghayeghi N, Sattler M, Salgia R, Griffin JD, Golemis EA, Freedman AS (February 1997). "Involvement of p130(Cas) and p105(HEF1), a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells". J. Biol. Chem. 272 (7): 4230–6. doi:10.1074/jbc.272.7.4230. PMID 9020138.
- Kyono WT, de Jong R, Park RK, Liu Y, Heisterkamp N, Groffen J, Durden DL (November 1998). "Differential interaction of Crkl with Cbl or C3G, Hef-1, and gamma subunit immunoreceptor tyrosine-based activation motif in signaling of myeloid high affinity Fc receptor for IgG (Fc gamma RI)". J. Immunol. 161 (10): 5555–63. PMID 9820532.
- Astier A, Manié SN, Law SF, Canty T, Haghayghi N, Druker BJ, Salgia R, Golemis EA, Freedman AS (December 1997). "Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in human B-cells: potential relevance to neoplastic lymphohematopoietic cells". Leuk. Lymphoma. 28 (1–2): 65–72. doi:10.3109/10428199709058332. PMID 9498705.
- Sattler M, Salgia R, Shrikhande G, Verma S, Uemura N, Law SF, Golemis EA, Griffin JD (May 1997). "Differential signaling after beta1 integrin ligation is mediated through binding of CRKL to p120(CBL) and p110(HEF1)". J. Biol. Chem. 272 (22): 14320–6. doi:10.1074/jbc.272.22.14320. PMID 9162067.
- Law SF, Zhang YZ, Fashena SJ, Toby G, Estojak J, Golemis EA (October 1999). "Dimerization of the docking/adaptor protein HEF1 via a carboxy-terminal helix-loop-helix domain". Exp. Cell Res. 252 (1): 224–35. doi:10.1006/excr.1999.4609. PMID 10502414.
- Suzuki T, Nakamoto T, Ogawa S, Seo S, Matsumura T, Tachibana K, Morimoto C, Hirai H (April 2002). "MICAL, a novel CasL interacting molecule, associates with vimentin". J. Biol. Chem. 277 (17): 14933–41. doi:10.1074/jbc.M111842200. PMID 11827972.
- Feng L, Guedes S, Wang T (July 2004). "Atrophin-1-interacting protein 4/human Itch is a ubiquitin E3 ligase for human enhancer of filamentation 1 in transforming growth factor-beta signaling pathways". J. Biol. Chem. 279 (28): 29681–90. doi:10.1074/jbc.M403221200. PMID 15051726.
- Liu X, Elia AE, Law SF, Golemis EA, Farley J, Wang T (December 2000). "A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member HEF1". EMBO J. 19 (24): 6759–69. doi:10.1093/emboj/19.24.6759. PMC 305889. PMID 11118211.